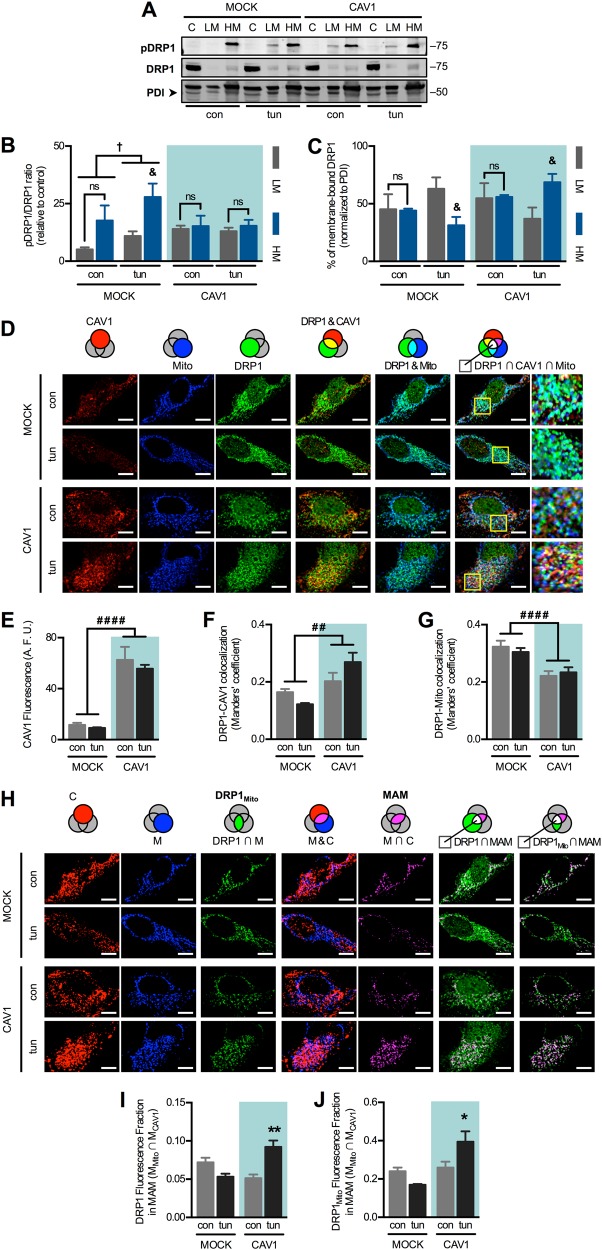
Fig. 6.
Caveolin-1 expression precludes DRP1 phosphorylation and redistribution during the early phase of ER stress. a Mock and CAV1 cells in control condition (con) or under early ER stress with tunicamycin (tun) were fractionated and processed for detection of phospho-DRP1 and DRP1 by western blotting using PDI as a loading control. C cytosol, LM light membranes (microsomes), HM heavy membranes (crude mitochondria). b Quantification of DRP1 phosphorylation analysed in (a) (n = 3). c Quantification of membrane-bound DRP1 distribution analysed in a (n = 3). d Experimental groups as in (a) were processed for immunofluorescence analysis. CAV1, mtHSP70 (mitochondrial marker, Mito) and DRP1 were stained with respective antibodies and then imaged using confocal microscopy. Merge between DRP1 and CAV1 images shows colocalized areas in yellow. Merge between DRP1 and Mito images shows colocalized areas in cyan. In merge images of Mito, DRP1 and CAV1, triple colocalization areas appear as white pixels. e CAV1 fluorescence per cell was quantified from images obtained in d (n = 3). f DRP1-CAV1 colocalization was quantified as Manders’ coefficients of images obtained in (d) (n = 3). g DRP1-Mito colocalization was quantified as Manders’ coefficients of images obtained in (d) (n = 3). h From images obtained in d, thresholded CAV1 (C) and Mito (M) images were generated. The intersection between M and DRP1 corresponds to mitochondrial DRP1 (DRP1mito). Merge between M and C images shows colocalized areas in magenta. The intersection between M and C corresponds to MAM. Merge between MAM and DRP1 or DRP1mito images shows colocalized pixels in white, which correspond to triple colocalization areas. i The fraction of DRP1 fluorescence that colocalizes with MAM was quantified from images obtained in h (n = 3). j The fraction of DRP1mito fluorescence that colocalizes with MAM was quantified from images obtained in h (n = 3). For each independent imaging experiment, 5–15 cells were analysed. Scale bars: 10 µm. Results are shown as mean ± s.e.m. &P < 0.05 compared with respective LM fraction. †P < 0.05 overall comparison between conditions. *P < 0.05 and **P < 0.05 compared with respective con condition. ##P < 0.01 and ####P < 0.0001 overall comparison between conditions. ns non-significant