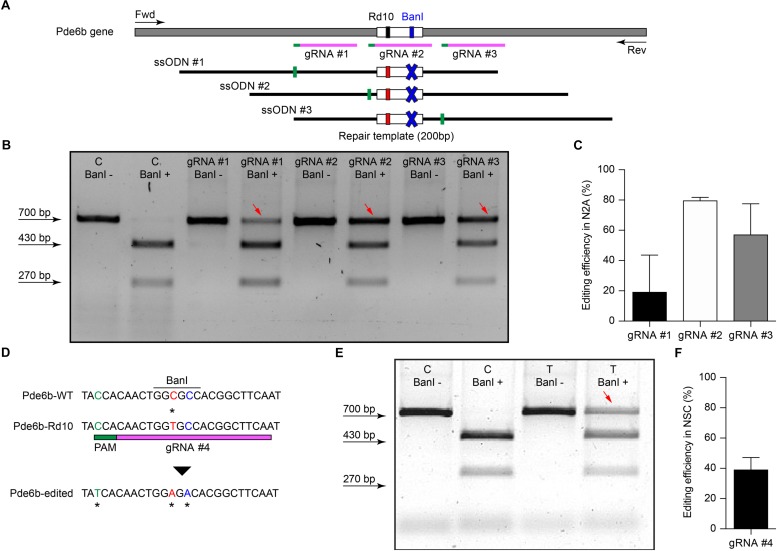
FIGURE 1.
Screening of gRNAs targeting the Rd10 locus. (A) Schematic representation (not in scale) of the mouse Pde6b gene showing the position of the three gRNAs tested (in magenta with green PAM sequence), the ssODN repair templates (black), and the PCR primers used for screening (arrows). The white rectangle represents the target editing region with Rd10 mutation (black/red) and the BanI cutting site (blue). Each ssODN also carries a silent mutation in the corresponding gRNA PAM sequence (green). (B) Representative example of an agarose gel electrophoresis of the BanI restriction assay from transfected (T) and control (C) mouse N2A cells. Unedited DNA is cut in two fragments by BanI digestion (470 and 230 bp), while edited DNA is not cut by the restriction enzyme (700 bp band, red arrows). (C) Quantification of the mean (±SD, n = 2) editing efficiency for the three gRNA in N2A cells. (D) Schematic representation of editing strategy for gRNA #4 targeting the Rd10 mutation. The HDR strategy was designed to edit the DNA sequence (in red), while introducing a silent mutation in the cutting sequence for BanI (in blue). A second silent mutation in the PAM sequence of the gRNA (in green) is included in the repair template in order to avoid further Cas9-mediated cutting on the edited genomic sequence. (E) Representative example of an agarose gel electrophoresis of the BanI restriction assay for gRNA #4 transfected (T) and control (C) NSC from Rd10 mice. The red arrow indicates the edited DNA that is resistant to BanI digestion. (F) Quantification of the mean (±SD, n = 3) editing efficiency for gRNA #4 in Rd10 NSC.