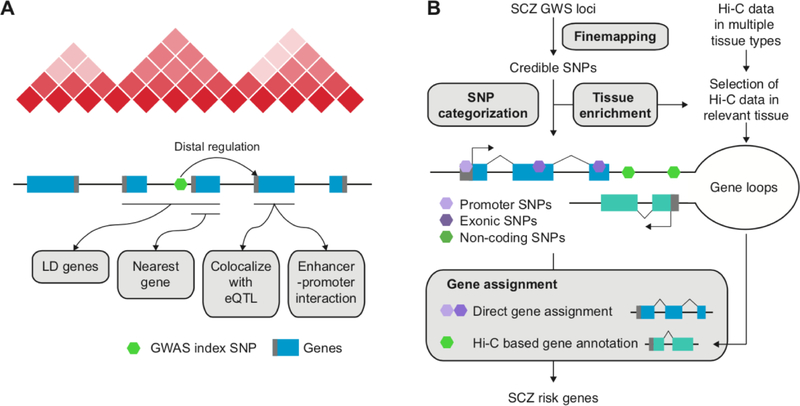
Figure 1. Defining putative target genes of genome-wide significant (GWS) loci for schizophrenia (SCZ).
A. Four widely used methods to identify target genes for a GWS locus. The nearest gene approach is to assign an index SNP to its nearest gene. The LD gene approach is to expand the search space to the LD region to include all genes located within the LD. Two methods leverage distal regulatory information such as eQTL and enhancer-promoter interactions. Enhancer-promoter interactions can be experimentally validated by Hi-C or computationally predicted. B. GWS loci for schizophrenia are finemapped to credible SNPs, which are subsequently categorized into promoter/exonic SNPs and non-coding SNPs. Promoter/exonic SNPs are directly assigned to their target genes, while non-coding SNPs are assigned to their target genes based on chromatin interaction profiles (Hi-C). The enrichment patterns of credible SNPs in regulatory regions of different tissue and cell types can inform us about which Hi-C dataset should be used to identify correct target genes.