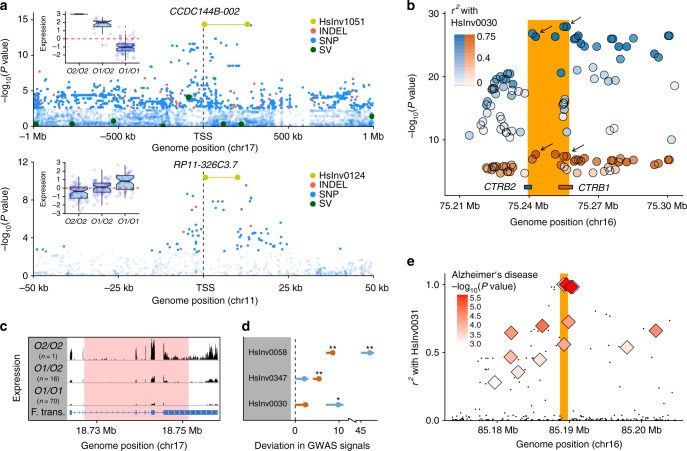
Fig. 5.
Examples of inversion functional effects. a Manhattan plots of logarithm-transformed linear regression t-test P values for cis-eQTL associations in LCLs of transcript CCDC144B-002 and gene RP11-326C3.7, showing inversions HsInv1051 (top) and HsInv0124 (bottom) as lead variants, together with boxplots of rank normalized expression and inversion genotype (centre, median; bounds of box, first and third quartiles; whiskers, smallest/largest value within 1.5 interquartile range; and notches, 95% confidence interval of the median). b Manhattan plot of logarithm-transformed GTEx P values of pancreas eQTLs for CTRB1 (red) and CTRB2 (blue) mapping around the HsInv0030 region (orange bar). Top lead eQTLs for each gene were the variants in highest LD with the inversion (rs9928842, r2 = 0.75; rs8057145, r2 = 0.73; black arrows). c Schematic diagram of average RNA-Seq expression profile in HsInv1051 genotypes of GEUVADIS LCL reads mapped to the inverted allele, showing the generation of a fusion transcript (F. trans.) with new 3′ sequences that loses the last two thirds of the gene exons. Inversion breakpoint is indicated in pink. d GWAS Catalog (orange) and GWASdb (blue) signals enrichment within individual inversions and flanking regions (±20 kb). Dot represents the mean and error bars the 0–0.95 confidence interval of the difference between the observed number of GWAS signals and a null distribution from 1000 random genomic regions for each inversion. One-tailed empirical test P value: **P < 0.01; *P < 0.05. e LD (r2) between HsInv0031 (orange bar) and 1000GP variants (black points) or Alzheimer’s disease GWAS signals (diamonds)78. Source data are provided as a Source Data file