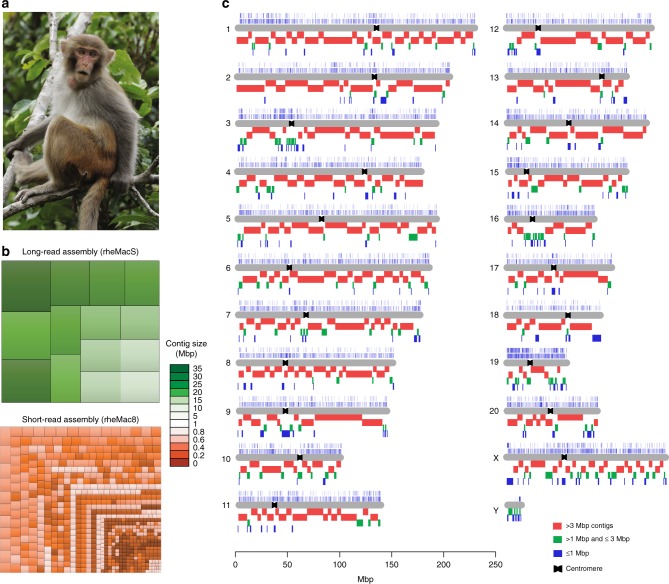
Fig. 1.
Long-read assembly of the Chinese rhesus macaque genome. a Chinese rhesus macaque (Macaca mulatta) (photograph courtesy by Jiaxin Zhao). b Treemaps for fragmentation difference between long-read and short-read rhesus assemblies. The rectangles represent the largest contigs that account for ~ 300 Mbp (~ 10%) of the assembly. c The chromosomal distribution of contigs of the rheMacS genome assembly. We compare genome sequence contiguity between rheMac8 and rheMacS. Thousands of gaps in rheMac8 were closed by longer contigs from rheMacS. The assembled contigs include > 3 Mbp (red), between 1 Mbp and 3 Mbp (green), and those < 1 Mbp (blue). The small contigs (< 1 Mbp) tend to consist of either centromeric or telomeric sequences. The centromeres of each chromosome are indicated based on previous annotation30