Abstract
Purpose: We aimed to evaluate the impact of HPV-driven carcinogenesis on outcome in vulvar squamous cell carcinoma patients (VSCC) treated with radiotherapy.
Methods and Materials: Analysis of clinical, pathological, and treatment data, HPV DNA-detection and -genotyping as well as p16INK4a immunohistochemistry were performed in 75 VSCC patients. Kaplan–Meier-method was used to estimate locoregional control (LC), Progression-free survival (PFS), and Overall Survival (OS). Univariate survival time comparisons were performed using the log-rank-test. Chi-square/Fisher exact test was used to assess correlations between HPV DNA and p16INK4a data, pathological, clinical, and treatment characteristics.
Results: 23/75 (30.67%) of all women had locoregional relapse, 7/75 (9.3%) systemic recurrence, and 35/75 (46.67%) died after a median follow-up of 26.4 months. 21.3% of the tumors were HPV DNA-positive, mostly (93.75%) for the high-risk (HR) HPV type 16. 25.3% showed p16INK4a-overexpression. 17.3% showed concomitant HPV DNA- and p16INK4a-positivity (cHPPVC). Patients with p16INK4a-overexpression, irrespective of the HPV DNA status, showed significantly better PFS (5-year-PFS 69.3 vs. 39.2%, p = 0.045), LC (5-year-LC 86.7 vs. 56.7%, p = 0.033) and a strong trend for better OS (5-year-OS 75.6 vs. 43.9%, p = 0.077). Patients with cHPPVC showed a trend for better PFS (5-year-PFS 72.7 vs. 41.3%, p = 0.082) and OS (5-year-OS 81.1 vs. 45.7%, p = 0.084) but no significant benefit for LC.
Conclusions: Patients with cHPPVC, indicating an etiological relevance of HPV in the respective tumors, showed a better, albeit not significant, prognosis. The sole detection of p16INK4a-overexpression is a prognostic factor for survival in vulvar cancer and indicates better prognosis after radiotherapy, independent of detection of HPV DNA. p16INK4a should be used as surrogate marker for HPV-driven carcinogenesis in vulvar cancer with caution.
Keywords: vulvar squamous cell carcinoma, human papillomavirus, p16INK4a, radiotherapy, prognostic factors
Introduction
Vulvar cancer represents only 3–5% of all gynecologic malignancies and prospective data regarding prognostic factors, outcome, and the role of HPV are rare. Two major etiologies have been described: HPV infection and chronic inflammatory dermatosis or autoimmune conditions. Histologically, precursor lesions of non-HPV-related vulvar carcinomas are differentiated vulvar intraepithelial neoplasia (VIN) whereas HPV-induced carcinomas arise from the usual type VIN and are of basaloid or warty type. HPV DNA is detected in >80% of all VIN lesions while HPV prevalence amongst invasive vulvar carcinomas seems to be lower. A recent publication (1) revealed that HPV contribution in invasive vulvar carcinoma worldwide has probably been overestimated: only 25.1% of 1,709 tumors from 39 countries were HPV-driven, as indicated by the simultaneous detection of HPV DNA and p16INK4a-overexpression. Among these, high-risk HPV 16 was the most common genotype (72.5%), followed by HPV 33 (6.5%) and HPV 18 (4.6%).
In HPV-transformed cells, expression of the viral oncogenes E6 and E7 leads to degradation and inactivation of the tumor suppressor proteins p53 and pRb, resulting in cell cycle dysregulation and uncontrolled cellular proliferation (2–4). E7 oncogene signaling further results in extensive overexpression of the cell cycle regulator p16INK4a, which is triggered by the removal of repressive trimethyl marks in the promoter region of the p16INK4a-encoding gene CDKN2A via the KDM6B demethylase (5). Since HPV DNA OR p16INK4a-overexpression may occur individually in different biological contexts, it is essential to assess the presence of both markers when determining a functional relevance of HPV in carcinogenesis (6). Only tumors, in which both—HR-HPV DNA AND diffuse p16INK4a-expression—are found, can biologically be considered as HPV-induced/HPV-driven (7).
The prognostic significance of the tumoral HPV status and the use of immunohistochemical p16INK4a-overexpression as a surrogate marker of HPV-induced transformation in vulvar squamous cell carcinoma (VSCC) are discussed controversially. Other squamous cell carcinomas, especially of the anogenital or head and neck region, are well-known to be associated with high-risk HPV. Furthermore, overexpression of the cell cycle regulator protein p16INK4a correlates with the presence of HPV DNA in cervical, anal or oropharyngeal cancer and p16INK4a-overexpression has been found to be of independent prognostic value for the response to radiation treatment (6, 8–13). A recent study revealed substantial mismatch between p16INK4a-overexpression and HPV DNA detection in VSCC and evidence arises that p16INK4a itself might function as an independent prognostic marker in vulvar cancer patients irrespective of an association with HPV (14, 15).
The study aimed to evaluate the association of HPV-driven carcinogenesis, indicated by HPV DNA and p16INK4a-overexpression, with clinical outcome in patients with VSCC treated with radiotherapy. In addition, the prognostic value of both markers was assessed individually in this cohort.
Materials and Methods
Patient Selection and Data Collection
Data from 119 patients with histologically proven vulvar squamous cell carcinoma who were treated with curatively intended radio- or radiochemotherapy at the Department of Radiation Oncology at University Hospital Heidelberg from 01/1999 to 04/2014 were retrieved from clinical databases. Tumor tissue biopsies from 92 patients taken at the time of primary diagnosis and/or time of disease recurrence were available for HPV DNA and p16INK4a analyses. The biopsies were obtained as formalin-fixed, paraffin-embedded tissue from local pathologists and the Institute of Pathology of University Hospital Heidelberg. For statistical analyses, only patients with tumor tissue samples and thus available HPV DNA- and p16INK4a-status from time of radiotherapy were included in the current analysis. Patient characteristics, tumor stage, details of oncological treatment including radiotherapy admission and follow-up exams were obtained from medical records. After exclusion of patients with insufficient clinical information, follow-up data or missing tumor tissue biopsies, 75 patients were included in the current analysis. Pathological and treatment characteristics are summarized in Tables 1, 2.
Table 1.
Clinical and pathological characteristics.
| Entire cohort n(75) | cHPPVC n(13)/17.3% | Non-cHPPVC n(62)/82.67% | p-value | p16INK4a positive n(19)/25.3% | p16INK4a negative n(56)/74.67% | p-value | |
|---|---|---|---|---|---|---|---|
| Date of first diagnosis | 0.139 | 0.058 | |||||
| 1991–1998 | 7 | 1 | 6 | 1 | 6 | ||
| 1999–2006 | 22 | 1 | 21 | 2 | 20 | ||
| 2007–2014 | 46 | 11 | 35 | 16 | 30 | ||
| T-status | 0.001* | 0.003* | |||||
| T1 | 35 | 6 | 29 | 10 | 25 | ||
| T2 | 29 | 1 | 28 | 2 | 27 | ||
| T3 | 7 | 4 | 3 | 4 | 3 | ||
| T4 | 3 | 2 | 1 | 3 | 0 | ||
| N-status at time of fist diagnosis | 0.564 | 0.420 | |||||
| N0 | 36 | 6 | 30 | 10 | 26 | ||
| N+ | 39 | 7 | 32 | 9 | 30 | ||
| ECS | 10 | 2 | 6 | 2 | 6 | ||
| N-status at time of first recurrence | 0.280 | 0.547 | |||||
| N0 | 44 | 8 | 36 | 11 | 33 | ||
| N+ | 22 | 2 | 20 | 5 | 17 | ||
| Grading | 0.225 | 0.423 | |||||
| G1 | 5 | 0 | 5 | 0 | 5 | ||
| G2 | 46 | 6 | 40 | 11 | 3235 | ||
| G3 | 20 | 6 | 14 | 7 | 13 | ||
| ≥ 8 mm | 17 | 3 | 14 | 5 | 11 | ||
| p16INK4a-status | <0.001* | - | |||||
| Positive | 19 | 13 | 6 | 19 | 0 | ||
| Negative | 56 | 0 | 56 | 0 | 56 | ||
| HPV-status | <0.001* | <0.001* | |||||
| Positive | 16 | 13 | 3 | 13 | 3 | ||
| Negative | 59 | 0 | 59 | 6 | 53 | ||
| Survival data | |||||||
| Deaths | 35 | 3 | 32 | 0.056 | 5 | 30 | 0.035* |
| Locoregional recurrence after RT | 23 | 2 | 21 | 0.163 | 2 | 21 | 0.023* |
| Distant metastases | 7 | 0 | 7 | 0.248 | 1 | 6 | 0.428 |
p <0.05, statistically significant.
Table 2.
Treatment characteristics.
| Entire cohort n(75) | cHPPVC n(13)/17.3% | non-cHPPVC n(62)/82.67% | p-value | p16INK4a positive n(19)/25.3% | p16INK4a negative n(56)/74.67% | p-value | |
|---|---|---|---|---|---|---|---|
| Surgical resection | 0.319 | 0.0445 | |||||
| Yes | 73 | 12 | 61 | 18 | 55 | ||
| No | 2 | 1 | 1 | 1 | 1 | ||
| Inguinal LNE | 0.511 | 0.205 | |||||
| Yes | 54 | 10 | 44 | 12 | 42 | ||
| No | 20 | 3 | 17 | 7 | 13 | ||
| Pelvic LNE | 0.045* | 0.165 | |||||
| Yes | 9 | 4 | 5 | 4 | 5 | ||
| No | 65 | 9 | 56 | 15 | 50 | ||
| R-status | 0.291 | 0.516 | |||||
| R0 | 61 | 10 | 51 | 16 | 45 | ||
| R+ | 6 | 2 | 4 | 2 | 4 | ||
| Resection margins | 0.588 | 0.551 | |||||
| <8 mm | 35 | 6 | 29 | 9 | 26 | ||
| ≥ 8 mm | 14 | 2 | 12 | 4 | 10 | ||
| Radiotherapy (RT) setting | 0.273 | 0.167 | |||||
| •Definitive RT | 5 | 2 | 3 | 2 | 3 | ||
| •Neoadjuvant RT | 3 | 1 | 2 | 2 | 1 | ||
| •Adjuvant RT | 67 | 10 | 57 | 15 | 52 | ||
| Radiotherapy timing | 0.006* | 0.033* | |||||
| RT as part of initial treatment | 44 | 12 | 32 | 15 | 29 | ||
| RT as part of salvage treatment | 31 | 1 | 30 | 4 | 27 | ||
| BED | |||||||
| >70 Gy | 18 | 4 | 14 | 4 | 14 | ||
| >60 Gy | 55 | 8 | 47 | 12 | 43 | ||
| >50 Gy | 72 | 13 | 59 | 19 | 53 |
p <0.05, statistically significant.
HPV DNA Detection and Genotyping
DNA extraction was performed using QIAGEN DNeasy© Blood and Tissue Kit from the available, formalin-fixed and paraffin-embedded tissue sections. For polymerase chain reaction (PCR) consensus HPV primers-sets (modified GP5+6+) were used. Samples with bands of about 150 base pairs corresponding to the length of positive control amplicons were considered positive.
We performed genotyping with positive samples using the Multiplex HPV Genotyping Kit© from Multimetrix GmbH (Regensburg, Germany, now DiaMex, Heidelberg, Germany) for subtypes HPV6, HPV11, HPV16, HPV18, HPV26, HPV31, HPV33, HPV35, HPV39, HPV42, HPV43, HPV44, HPV45, HPV51, HPV52, HPV53, HPV56, HPV58, HPV59, HPV66, HPV68, HPV70, HPV73, and HPV82. Samples were considered positive if median intensity of fluorescence in the Luminex Analyzer was 10 or higher.
p16INK4a Immunohistochemistry
p16INK4a immunohistochemistry was performed on 2 μm tissue sections using the CINtec p16INK4a histology kit (mtm Laboratories, Heidelberg, Germany) according to the manufacturer's instructions. The p16INK4a antigen was detected with a mouse monoclonal anti-human p16INK4a antibody (E6H4™). Tissue sections with a diffuse (clonal) tumoral p16INK4a staining as previously described (10) in more than 50% of tumor cells were considered positive. Lesions showing either focal staining only or no p16INK4a expression at all were considered as p16INK4a-negative.
Statistical Analysis
All survival end-points were calculated starting from the date of start of radiotherapy. Overall survival (OS) was then defined as time to death from any cause. Time to locally progressive disease of the primary tumor or regional lymph nodes was determined as locoregional control (LC). Progression-free survival (PFS) was defined as time to local/distant recurrence or death. Distant control (DC) was defined as time to distant metastases. All patients with no event at the last follow-up were censored. In detail, events were death for OS, locoregional progressive disease, distant metastases or death for PFS, local recurrence or recurrence in regional lymph nodes for LC, distant metastases for DC. Chi-square/Fisher exact test were used to assess correlations between staining results and pathological, clinical, and treatment characteristics. Differences between the p16INK4a-positive vs. the p16INK4a-negative group as well as the group with cHPPVC (concomitant HPV and P16INK4a Positive Vulvar Cancer) vs. the rest of the patients were assessed using the chi square/Fisher exact test. The Kaplan–Meier method was used to estimate LC, PFS, DC, and OS for various group partitions. Univariate survival time comparisons were performed using the log-rank test. The statistical analysis was performed using SPSS version 24. The study was granted ethical approval by the local ethics committee.
Results
Patients' and Treatment Characteristics (Tables 1, 2)
At the time of primary diagnosis median age was 68 years (range 37–88 years). 62 women were diagnosed with a tumor ≤ T2 (82.7%), 39 women presented with nodal involvement (52%) at time of first diagnosis and 22 at time of first recurrence (29.3%) (Table 1). All in all, 21 patients (28%) received radiotherapy to the vulva only, 18 patients (24%) received radiotherapy to the lymphatic drainage (inguinal and/or iliacal) only and 36 patients (48%) received radiotherapy to both vulva and lymphatic drainage. All patients received radiotherapy with curative intent with radiation doses > 45 Gy BED (biological effective dose, calculated with an alpha/beta of 10). Mean BED was 63.7 Gy (range 46.7–75.15 Gy). 72 patients received more than 50 Gy BED, 55 received more than 60 Gy BED, and 18 received more than 70 Gy BED (Table 2). Median follow-up of the entire cohort was 26.4 months (range 2.4–160.3 months, counted from the start of radiotherapy). 23 patients had locoregional relapse (30.7%), 7 developed systemic recurrence (9.3%), and 35 died (46.7%).
44 patients underwent radio-/radiochemotherapy as part of the initial treatment, 40 of them as adjuvant treatment after resection, three in a neoadjuvant setting and one as definitive treatment. 6 patients received concomitant chemotherapy [MitomycinC/5-FU (n = 5) or Cisplatin weekly (n = 1)]. After a median follow-up of 28.3 months (range 2.4–128.3 months) of this subgroup, 8 patients had locoregional relapse (18%), 5 patients developed systemic recurrence (11.4%), and 15 patients died (34%).
31 patients received radiotherapy because of disease recurrence, 27 of them as adjuvant treatment after resection, and 4 as definitive treatment. After a median follow-up of 14.4 months (range 2.5–160.3 months), 15 patients had locoregional relapse after radiotherapy (48.4%), two developed systemic recurrence (6.5%) and 20 had died (64.5%).
Results of p16INK4a Immunohistochemistry and HPV DNA Genotyping (Table 3)
Table 3.
Results of p16INK4a-immunostaining and expression of HPV DNA.
| p16INK4a | Total | |||
|---|---|---|---|---|
| Negative | Positive | |||
| HPV DNA | Negative | 53 (70.67%) | 6 (8%) | 59 (78.67%) |
| Positive | 3 (4%) | 13 (17.3%)* | 16 (21.3%) | |
| Total | 56 (74.67%) | 19 (25.3%) | 75 (100%) | |
cHPPVC.
Tissue specimen from time of primary diagnosis and/or tumor specimen from disease recurrence were available for analysis. For all 44 patients who underwent radio-/radiochemotherapy as part of the initial treatment tissue specimen from time of primary diagnosis were available for analysis. For all 31 patients who received radiotherapy because of disease recurrence, tissue specimen were available from diagnosis of recurrence. From this subgroup, additional tissue specimens from time of initial diagnosis were available for 13 patients. Interestingly, one patient initially tested positive for HPV DNA but without p16INK4a-overexpression did not show HPV DNA in tissue specimens of disease recurrence. Two women initially showing no p16INK4a-overexpression and no HPV DNA showed p16INK4a-overexpression at recurrence. For statistical analyses, HPV DNA- and p16INK4a-status at time of radiotherapy were used.
59/75 (78.7%) of VSCC were tested HPV DNA-negative, whereas HR-HPV DNA was detected in 16/75 (21.3%) of the tumors. HPV 16 was the most frequently detected genotype (15/16, 93.75%), only one tumor specimen was positive for HPV 33/59. 19/75 patients (25.3%) showed p16INK4a-overexpression, defined as diffuse staining in more than 50% of tumor cells; 56/75 patients (74.67%) were tested p16INK4a-negative, showing only focal or no staining at all. 53/86 patients (70.67%) were both HPV DNA and p16INK4a-negative, while 13/75 (17.3%) were tested positive for both parameters. 6/75 patients (8%) were tested p16INK4a-positive without detection of HPV DNA. 3/75 patients (4%) were HPV DNA-positive without showing p16INK4a-overexpression (Table 3).
Chi-Square/Fisher exact test revealed significant correlation between detection of HPV DNA and p16INK4a-overexpression (p <0.001). Furthermore, a significant correlation between cHPPVC and tumor stage (p = 0.001) and p16INK4a -status and tumor stage (p = 0.003) could be observed. Interestingly, cHPPVC and p16INK4a-overexpression were associated with higher tumor stage (>T2). Chi-Square/Fisher exact test revealed no further correlations of cHPPVC or p16INK4a-overexpression with any of the assessed pathological, patients' or treatment characteristics like age, date of primary diagnosis, nodal status, extracapsular tumor spread, grading, lymphovascular space invasion, radiation dose or setting of radiotherapy (adjuvant vs. definitive vs. neoadjuvant) (see also Tables 2, 3).
Survival Endpoints for the Entire Cohort
Kaplan-Meier-estimated median PFS of the entire cohort was 28 months (95%-CI 0-77.4 months) with 2- and 5-year-PFS rates of 51.7 and 46.4%, respectively. Estimated median LC had not been reached at the time of analysis. 2- and 5-year LC rates were 69.2 and 64.1%, respectively. In the entire cohort, seven patients (9.3%) showed systemic recurrence. Kaplan-Meier estimated 1- and 2-year DC rates were 91.4 and 89.4%, respectively. All seven patients were in the non-cHPPVC group, so that further statistical analyses of DC were not reasonable. Estimated median OS was 66.4 months with 2-, 5-, and 10-year-OS rates of 58.9, 51.5, and 45.9%, respectively.
Survival Endpoints by p16 INK4a-Status Alone
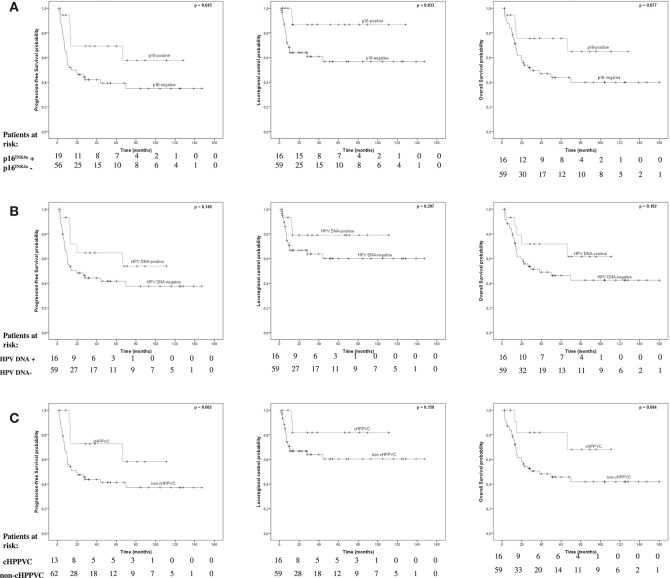
p16INK4a-overexpression was associated with significantly better PFS and LC rates and a strong trend for a better OS (Figure 1). Kaplan-Meier-estimated median PFS, LC, and OS for p16INK4a-positive patients had not been reached at time of analysis. For p16INK4a-negative patients median PFS and OS were 14.1 months (95%-CI 0-32.9 months) and 29.3 months (95%-CI 0-61.6 months), respectively, estimated median LC had not been reached, yet. Patients with p16INK4a-overexpression showed significantly better PFS (p = 0.045) with 5-year-PFS rates of 69.3 vs. 39.2% for p16INK4a-negative patients. Patients with p16INK4a-overexpression showed significantly better LC rates (p = 0.033) with 5-year LC rates of 86.7 vs. 56.7% for p16INK4a-negative patients. p16INK4a-overexpression was associated with better, albeit not significant, OS (p = 0.077) with 5-year-OS rates of 75.6 vs. 43.9% for p16INK4a-negative patients.
Figure 1.
Survival endpoints by, p16INK4a-, HPV-, and cHPPVC-status. Kaplan-Meier estimated Progression-free survival (PFS), Locoregional Control (LC), and Overall Survival (OS) stratified by p16INK4a-status (A), HPV DNA-status (B), and cHPPVC-status (C) at time of radiotherapy.
Survival Endpoints by HPV DNA-Status Alone
Detection of HPV DNA alone was not associated with significantly better PFS, LC or OS rates (Figure 1). Kaplan-Meier-estimated median PFS for HPV-DNA-positive patients had not been reached at time of analysis and was 19.4 months (95%-CI 0-39.7 months) for HPV-DNA-negative patients. There were no significant differences regarding PFS (p = 0.14) with 5-year-PFS rates of 64.6% for HPV DNA-positive patients vs. 41.7% for HPV DNA-negative patients. Estimated median LC for both HPV DNA-positive and -negative patients had not been reached at time of analysis. There were no significant differences regarding LC (p = 0.207) with 5-year-LC rates of 79% for HPV DNA-positive patients vs. 60% for HPV DNA-negative patients. Estimated median OS for HPV DNA-positive patients had also not been reached yet, and was 38.3 months for HPV DNA-negative patients (95%-CI 0-87.2 months). There were no significant differences regarding OS (p = 0.182) with 5-year-OS rates of 71.8% for HPV DNA-positive patients vs. 46.3% for HPV DNA-negative patients.
Survival Endpoints by cHPPVC-Status
cHPPVC-status was associated with a strong trend for better PFS and OS rates, with no significant differences in LC rates (Figure 1). Kaplan-Meier-estimated median PFS for cHPPVC patients had not been reached at time of analysis and was 19.4 months (95%-CI 1.4-37.4 months) for non-cHPPVC patients. Patients with cHPPVC showed a strong trend for better PFS (p = 0.082) with 5-year-PFS rates of 72.7 vs. 41.3% for non-HPPVC patients. Estimated median LC for both cHPPVC and non-cHPPVC patients had not been reached at time of analysis. There were no significant differences regarding LC (p = 0.158) for patients with cHPPVC compared to non-cHPPVC patients with 5-y-LC rates of 81.8 vs. 60.4%. Estimated median OS for cHPPVC patients had also not been reached yet, and was 38.3 months for non-HPPVC patients (95%-CI 0-82.4 months). Patients with cHPPVC showed a strong trend for better OS (p = 0.084) with 5-year-OS rates of 81.1 vs. 45.7% for non-HPPVC patients.
Discussion
Data on the prognostic significance of HPV-driven carcinogenesis in vulvar cancer are discussed controversially. The main challenge in comparing literature are differences in defining HPV-driven carcinogenesis. Biologically, three criteria are necessary to prove a causative role of HPV in carcinogenesis: presence of HPV DNA, transcription and translation of the E6 and E7 oncogenes and dependence of HPV-transformed cells on their continuous expression (6). Due to difficulties in defining such criteria in daily clinical practice, practicable markers indicating a functional, transforming relevance of HPV in tumor cells are needed. In this context, p16INK4a has become a widely used biomarker for HPV-transformed cells (6, 16), e.g., in the anogenital and head and neck region (6, 10–12).
In our cohort, concomitant p16INK4a-overexpression and detection of HPV DNA was associated with a trend for better PFS and OS, whereas p16INK4a-overexpression alone was associated with significantly better PFS and LC and a strong trend for a better OS. Sole detection of HPV DNA was not associated with significantly better PFS, LC, or OS rates.
Several studies were published assessing the prognostic role of HPV-driven carcinogenesis in vulvar cancer, most of them using either p16INK4a-overexpression (17–20) or HPV DNA (17, 18, 21–23) as marker for HPV-driven carcinogenesis. They couldn't prove an independent prognostic role of HPV DNA in vulvar cancer (21, 22) or solely claimed a trend for better survival (23). Interestingly, in studies using p16INK4a-overexpression only as surrogate marker, a survival benefit has been reported (17–20). A recently published study describes a survival benefit for HPV-driven VSCC patients undergoing primary resection (24). VSCCs were considered HPV-related in case of either >25% p16INK4a-expression and HPV-positivity or >25% p16INK4a-expression and high grade squamous intraepithelial lesion next to the tumor without HPV-positivity (24). It has been shown that precursor lesions of vulvar cancer are more often HPV-associated than invasive VSCCs (>80 vs. 25%) (1), so that HPV-driven etiology might be overestimated in that collective. Additionally, HPV-negative patients presented with positive lymph nodes more frequently, being strongly associated with a worse prognosis which might bias the reported results.
Our study reports one of the largest cohorts of irradiated VSCC patients with assessed HPV DNA and p16INK4a status. An outstanding characteristic of our study is that concomitant detection of HPV DNA and p16INK4a-overexpression was regarded mandatory for indicating HPV-driven carcinogenesis (7). As all patients required radiotherapy or already were in the situation of recurrence this is considered to be a negatively selected collective. Furthermore, data might be biased due to the retrospective nature of the study. As only 17.3% were identified having cHPPVC, the cohort might be too small to detect any statistically significant differences in oncological outcome. However, Alonso et al. also used HPV DNA in combination with p16INK4a-overexpression as marker for HPV-driven carcinogenesis. Only cases with diffuse staining, defined as continuous staining of cells of the basal and parabasal layers, were considered p16INK4a-positive. The authors also couldn't find significant differences in outcome for HPV-driven VSCC patients (25), what is consistent with our data. There were no anaylses conducted regarding the prognostic significance of p16INK4a-overexpression alone, what was associated with significantly better LC, PFS, and better OS in our cohort, irrespective of HPV DNA-detection.
Recently, more and more evidence arises revealing a mismatch between p16INK4a-overexpression and HPV DNA-detection in VSCCs. In our cohort, six patients were tested p16INK4a-positive but HPV DNA-negative, three were HPV DNA-positive but p16INK4a-negative. A large cohort study reported only 87.9% to be both HPV DNA- and p16INK4a-positive (1). Sznurkowsky et al. (15) described 29% of 85 tumors p16INK4a-positive but HPV DNA-negative and 24% p16INK4a-negative but HPV DNA-positive. Only p16INK4a-overexpression correlated with prolonged OS and predicted a better response to irradiation (15). Additionally, a large meta-analysis of 2,309 patients suggested that p16INK4a-status itself is associated with a better prognosis in vulvar cancer patients and indicates higher radiosensitivity (14). There are current studies investigating other molecular mechanisms leading to higher radiosensitivity of p16INK4a-positive VSCCs, suggesting that immunologic effects depending on p16INK4a-overexpression contribute to better outcome (26).
Patients with cHPPVC, indicating an etiological relevance of HPV in the respective tumors, showed a better, albeit not significant, prognosis. Due to the rather low incidence of cHPPVC, the sample size might be too small for showing any statistically significant differences in our cohort. Our observations should consequently be validated in larger patient cohorts. Interestingly, p16INK4a overexpression alone seems to be a prognostic factor for survival in vulvar cancer and indicates better prognosis after radiotherapy, independent of detection of HPV DNA. p16INK4a should be used as surrogate marker for HPV-driven carcinogenesis in vulvar cancer only with caution, as more and more evidence arises that there seem to be other HPV-independent mechanisms in vulvar cancer leading to p16INK4a overexpression. Further analyses are necessary investigating potential molecular mechanisms for p16INK4a overexpression in vulvar cancer.
Data Availability
The datasets for this manuscript are not publicly available due to privacy and data protection reasons of the patient data included in the analysis. Requests to access the datasets should be directed to NA, nathalie.arians@med.uni-heidelberg.de.
Ethics Statement
This study was carried out in accordance with the recommendations of the Declaration of Helsinki and institutional ethical policies of the University Hospital Heidelberg. The protocol was approved by the ethics committee of the University Hospital Heidelberg (S-308/2012, 24.09.2012, Amendment 04.06.2018). As the data were analyzed retrospectively and anonymously and treatment of patients was not affected by this study, no written informed consent from each individual patient was necessary according to institutional standards and the local ethics committee decision.
Author Contributions
NA, E-SP, MR, and KL: conceptualization. NA and TN: data curation and investigation. NA: formal analysis and writing—original draft. NA, E-SP, and MR: methodology and validation. JD, MK, and KL: project administration, resources, and supervision. E-SP and SK: writing—review and editing. All authors contributed substantially to the work reported.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jonathan Dörre for all the technical support in performing HPV DNA-detection and -genotyping as well as p16INK4a immunohistochemistry.
Footnotes
Funding. We acknowledge financial support by Deutsche Forschungsgemeinschaft within the funding programme Open Access Publishing, by the Baden-Württemberg Ministry of Science, Research and the Arts and by Ruprecht-Karls-Universität Heidelberg.
References
- 1.de Sanjose S, Alemany L, Ordi J, Tous S, Alejo M, Bigby SM, et al. Worldwide human papillomavirus genotype attribution in over 2000 cases of intraepithelial and invasive lesions of the vulva. Eur J Cancer. (2013) 49:3450–61. 10.1016/j.ejca.2013.06.033 [DOI] [PubMed] [Google Scholar]
- 2.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. (1990) 63:1129–36. 10.1016/0092-8674(90)90409-8 [DOI] [PubMed] [Google Scholar]
- 3.Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. (1989) 8:4099–105. 10.1002/j.1460-2075.1989.tb08594.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duensing S, Lee LY, Duensing A, Basile J, Piboonniyom S, Gonzalez S, et al. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci USA. (2000) 97:10002–7. 10.1073/pnas.170093297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaughlin-Drubin ME, Crum CP, Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci USA. (2011) 108:2130–5. 10.1073/pnas.1009933108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prigge ES, von Knebel Doeberitz M, Reuschenbach M. Clinical relevance and implications of HPV-induced neoplasia in different anatomical locations. Mutat Res Rev Mutat Res. (2017) 772:51–66. 10.1016/j.mrrev.2016.06.005 [DOI] [PubMed] [Google Scholar]
- 7.von Knebel Doeberitz M. The causal role of human papillomavirus infections in non-anogenital cancers. It's time to ask for the functional evidence. Int J Cancer. (2016) 139:9–11. 10.1002/ijc.30059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindel K, Burri P, Studer HU, Altermatt HJ, Greiner RH, Gruber G. Human papillomavirus status in advanced cervical cancer: predictive and prognostic significance for curative radiation treatment. Int J Gynecol Cancer. (2005) 15:278–84. 10.1111/j.1525-1438.2005.15216.x [DOI] [PubMed] [Google Scholar]
- 9.Lindel K, Beer KT, Laissue J, Greiner RH, Aebersold DM. Human papillomavirus positive squamous cell carcinoma of the oropharynx: a radiosensitive subgroup of head and neck carcinoma. Cancer. (2001) 92:805–13. [DOI] [PubMed] [Google Scholar]
- 10.Prigge E-S, Toth C, Dyckhoff G, Wagner S, Muller F, Wittekindt C, et al. p16(INK4a) /Ki-67 co-expression specifically identifies transformed cells in the head and neck region. Int J Cancer. (2015) 136:1589–99. 10.1002/ijc.29130 [DOI] [PubMed] [Google Scholar]
- 11.Smeets SJ, Hesselink AT, Speel E-JM, Haesevoets A, Snijders PJF, Pawlita M, et al. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int J Cancer. (2007) 121:2465–72. 10.1002/ijc.22980 [DOI] [PubMed] [Google Scholar]
- 12.Rietbergen MM, Leemans CR, Bloemena E, Heideman DAM, Braakhuis BJM, Hesselink AT, et al. Increasing prevalence rates of HPV attributable oropharyngeal squamous cell carcinomas in the Netherlands as assessed by a validated test algorithm. Int J Cancer. (2013) 132:1565–71. 10.1002/ijc.27821 [DOI] [PubMed] [Google Scholar]
- 13.Koerber SA, Schoneweg C, Slynko A, Krug D, Haefner MF, Herfarth K, et al. Influence of human papillomavirus and p16(INK4a) on treatment outcome of patients with anal cancer. Radiother Oncol. (2014) 113:331–6. 10.1016/j.radonc.2014.11.013 [DOI] [PubMed] [Google Scholar]
- 14.Cao H, Wang S, Zhang Z, Lou J. Prognostic value of overexpressed p16(INK4a) in vulvar cancer: a meta-analysis. PLoS ONE. (2016) 11:e0152459 10.1371/journal.pone.0152459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sznurkowski JJ, Zawrocki A, Biernat W. The overexpression of p16 is not a surrogate marker for high-risk human papilloma virus genotypes and predicts clinical outcomes for vulvar cancer. BMC Cancer. (2016) 16:465. 10.1186/s12885-016-2503-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergeron C, Ronco G, Reuschenbach M, Wentzensen N, Arbyn M, Stoler M, et al. The clinical impact of using p16(INK4a) immunochemistry in cervical histopathology and cytology: an update of recent developments. Int J Cancer. (2015) 136:2741–51. 10.1002/ijc.28900 [DOI] [PubMed] [Google Scholar]
- 17.Lee LJ, Howitt B, Catalano P, Tanaka C, Murphy R, Cimbak N, et al. Prognostic importance of human papillomavirus (HPV) and p16 positivity in squamous cell carcinoma of the vulva treated with radiotherapy. Gynecol Oncol. (2016) 142:293–8. 10.1016/j.ygyno.2016.05.019 [DOI] [PubMed] [Google Scholar]
- 18.Yap ML, Allo G, Cuartero J, Pintilie M, Kamel-Reid S, Murphy J, et al. Prognostic significance of human papilloma virus and p16 expression in patients with vulvar squamous cell carcinoma who received radiotherapy. Clin Oncol. (2018) 30:254–61. 10.1016/j.clon.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 19.Horne ZD, Dohopolski MJ, Pradhan D, Bhargava R, Edwards RP, Kelley JL, et al. Human papillomavirus infection mediates response and outcome of vulvar squamous cell carcinomas treated with radiation therapy. Gynecol Oncol. (2018) 151:96–101. 10.1016/j.ygyno.2018.08.002 [DOI] [PubMed] [Google Scholar]
- 20.Dohopolski MJ, Horne ZD, Pradhan D, Bhargava R, Edwards RP, Kelley JL, et al. The prognostic significance of p16 status in patients with vulvar cancer treated with vulvectomy and adjuvant radiation. Int J Radiat Oncol Biol Phys. (2018) 103:152–60. 10.1016/j.ijrobp.2018.08.014 [DOI] [PubMed] [Google Scholar]
- 21.Monk BJ, Burger RA, Lin F, Parham G, Vasilev SA, Wilczynski SP. Prognostic significance of human papillomavirus DNA in vulvar carcinoma. Obstet Gynecol. (1995) 85:709–15. 10.1016/0029-7844(95)00045-S [DOI] [PubMed] [Google Scholar]
- 22.Pinto AP, Schlecht NF, Pintos J, Kaiano J, Franco EL, Crum CP, et al. Prognostic significance of lymph node variables and human papillomavirus DNA in invasive vulvar carcinoma. Gynecol Oncol. (2004) 92:856–65. 10.1016/j.ygyno.2003.11.052 [DOI] [PubMed] [Google Scholar]
- 23.Kim Y, Kim J-Y, Kim JY, Lee NK, Kim JH, Kim YB, et al. Treatment outcomes of curative radiotherapy in patients with vulvar cancer: results of the retrospective KROG 1203 study. Radiat Oncol J. (2015) 33:198–206. 10.3857/roj.2015.33.3.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinten F, Molijn A, Eckhardt L, Massuger LFAG, Quint W, Bult P, et al. Vulvar cancer: two pathways with different localization and prognosis. Gynecol Oncol. (2018) 149:310–7. 10.1016/j.ygyno.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 25.Alonso I, Fuste V, del Pino M, Castillo P, Torne A, Fuste P, et al. Does human papillomavirus infection imply a different prognosis in vulvar squamous cell carcinoma? Gynecol Oncol. (2011) 122:509–14. 10.1016/j.ygyno.2011.05.016 [DOI] [PubMed] [Google Scholar]
- 26.Sznurkowski JJ, Zawrocki A, Biernat W. Local immune response depends on p16(INK4a) status of primary tumor in vulvar squamous cell carcinoma. Oncotarget. (2017) 8:46204–10. 10.18632/oncotarget.17581 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets for this manuscript are not publicly available due to privacy and data protection reasons of the patient data included in the analysis. Requests to access the datasets should be directed to NA, nathalie.arians@med.uni-heidelberg.de.