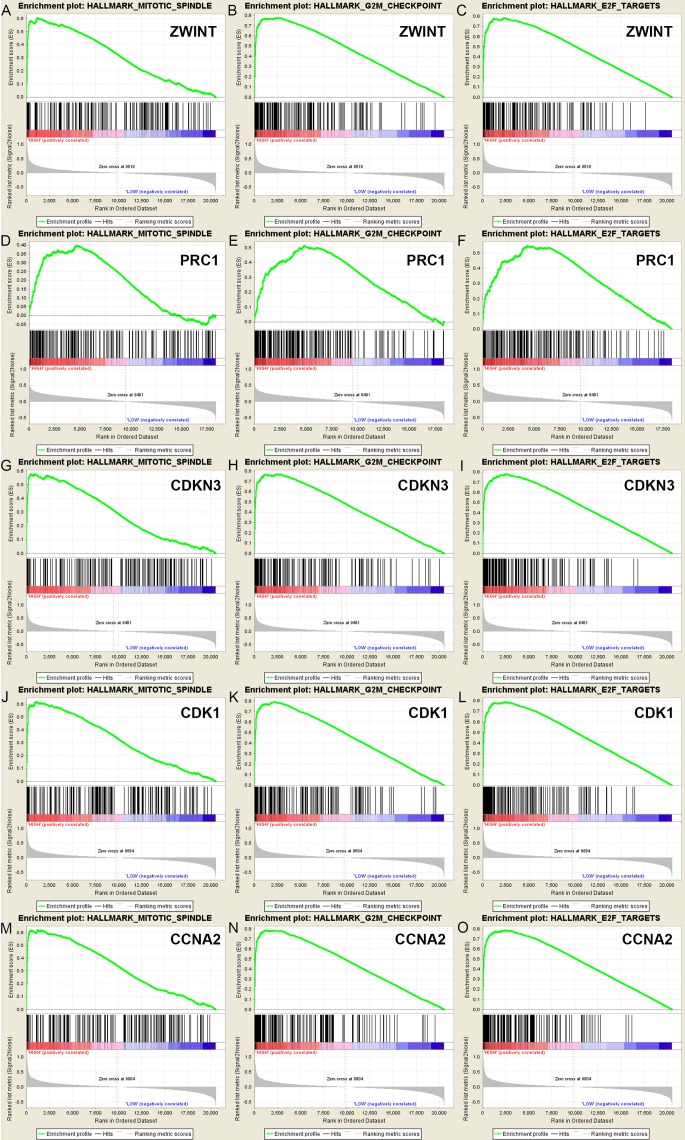
Figure 6.
A total of 100 significant genes were obtained from GSEA with positive and negative correlation. GSEA was used to perform hallmark analysis in ZWINT, PRC1, CDKN3, CDK1 and CCNA2, respectively. Results of GESA suggested that (A–C) ZWINT, (D–F) PRC1, (G–I) CDKN3, (J–L) CDK1, (M–O) CCNA2 significantly involved in the same hallmarks pathways including mitotic spindle, G2M checkpoint and E2F targets.