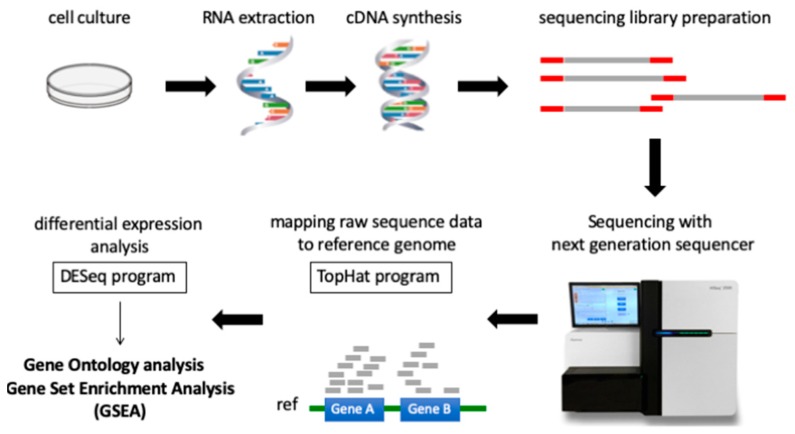
Figure 1.
The workflow of RNA-seq and data analysis used in this study. Next Generation Sequencing reads sequenced with the Illumina Hiseq 2500 were aligned to the hg19 genome + transcriptome assembly (UCSC hg19) using TopHat v2.1.1 with the default parameters. Read counts of each gene were obtained using HTSeq and differential expression was analyzed using DESeq. Gene Ontology (GO) analysis and Gene Set Enrichment Analysis (GSEA) were performed to interpret the results.