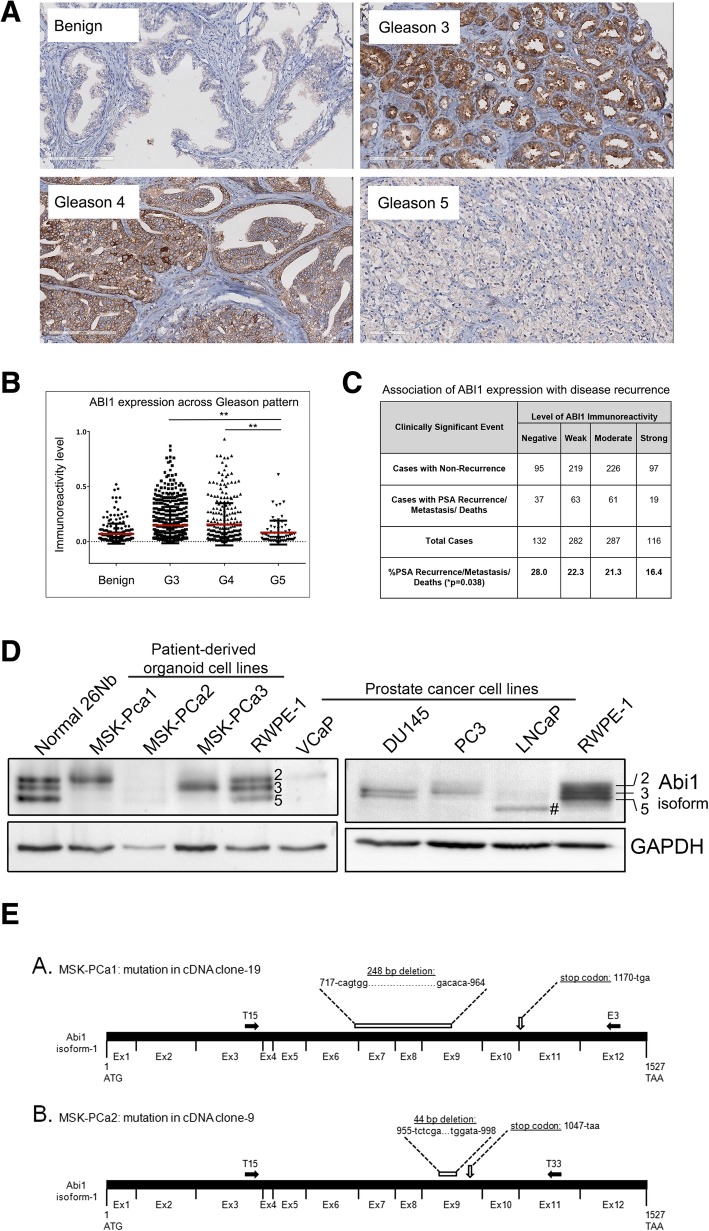
Fig. 1.
ABI1 expression is downregulated during prostate tumor progression. Prostate cancer tissue microarray (n = 505 patients) was stained for ABI1 to evaluate its expression during prostate tumor progression. (a) Representative ABI1 expression staining in Gleason 3, 4, and 5 patterns, as indicated; Benign indicates normal prostate epithelium. (b) Comparison of ABI1 expression among Gleason patterns, 3, 4 and 5 indicating significant downregulation of ABI1 in Gleason 5 vs. 3, (p = 0.0012); or 4 (p = 0.0025) (one-way ANOVA, multiple comparison). (c) Analysis of ABI1 expression in the tissue microarray indicating an association between low ABI1 expression and clinically significant events, such as biochemical recurrence, a metastatic event, and death; p = 0.038; 99% confidence interval: 0.03308–0.4292; Linear-by-Linear association exact test (Sytel Studio-9). (d) Representative Western blots showing reduced levels of ABI1 in different patient-derived organoid cell lines (left) and immortalized prostate cancer cell lines (right) compared with non-tumor 26Nb and RWPE-1 cells. Numbers next to bands in the RWPE-1 lane and on the right side of the panel indicate ABI1 isoform designation: top band, isoform 2; middle band, isoform 3; and bottom band, isoform 5 [34, 40]; #, indicates the mutant ABI1 lacking exon 6 in LNCaP [37]. GAPDH was used as the loading control. (e) Identification of mutations in the cDNA of ABI1 from organoid cultures of human metastatic prostate cancer cell lines. (A. and B.) Schematic of full-length ABI1 cDNA (isoform-1) with coding regions of consecutive exons (Ex1-Ex12) indicated (not drawn to scale). The ATG start codon (corresponding to nucleotide 1), TAA stop codon (corresponding to nucleotide 1527), and the T15, E3, and T33 primer sites (filled-in black arrows) are labeled. The T15, E3, and T33 primers [40] were used to selectively amplify ABI1 cDNA via PCR from the MSK-PCa1 and MSK-PCa2 organoid cell lines. PCR products were subcloned into a pCR-Blunt II TOPO vector and sequenced. A. Schematic of mutation identified in cDNA clone-19 sequence from MSK-PCa1 cells. Sequencing of cDNA clone-19 revealed a 248-bp deletion spanning nucleotides 717 to 964 (from the 3′ end of Exon6, at the junction between Exon 6 and Exon 7, to the middle of Exon 9) that is predicted to result in a frame shift and subsequent stop codon at nucleotide 1170. B. Schematic of mutation identified in cDNA clone-9 from MSK-PCa2 cells. Sequencing of cDNA clone-9 revealed a 44-bp deletion spanning nucleotides 955 to 998 in the middle of Exon 9 that is predicted to result in a frame shift and subsequent stop codon at nucleotide 1047