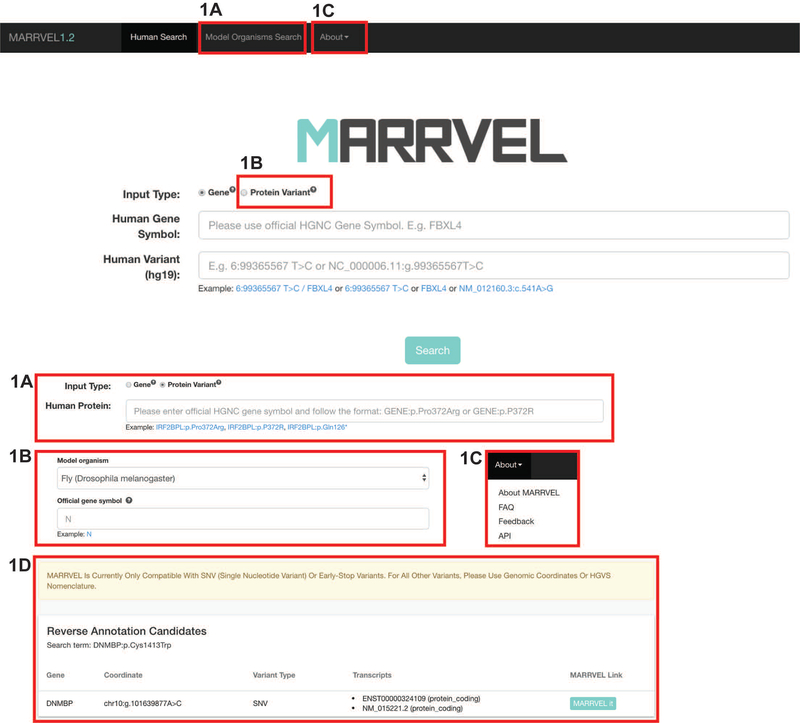
Figure 1: Annotated screenshot of search page.
A) Click on Model organism Search for starting a search based on a gene in a model organism. The user will be directed to a page to select the model organism of interest and then enter the gene symbol. Suggestions of gene symbols will be available as the user types.
B) For variants with amino acid/protein nomenclature, click on “Protein Variant”. Users will see a single search bar where both the gene and variant can be entered. The search bar accepts either three letter amino acid code or single letter amino acid code. When there is ambiguity for which isoform the user is referring to, all options will be listed for the user to select.
C) For “About MARRVEL”, “FAQ”, “Feedback”, and “API”, click on “About” on the top menu. “About MARRVEL” includes information on the development team, acknowledgements of data sources, and more. “FAQ” includes the answers to frequently asked questions. “Feedback” page allows users to report errors or provide suggestions for new features in future updates. “API” page provides instructions on how to access MARRVEL API.
D) After searching for DNMBP:p.Cys1413Trp from the “Protein Variant” search bar, a box titled “Reverse Annotation Candidates” displays the gene name, genomic coordinate of the variant, variant type, transcripts, and a link to proceed to the MARRVEL results page.