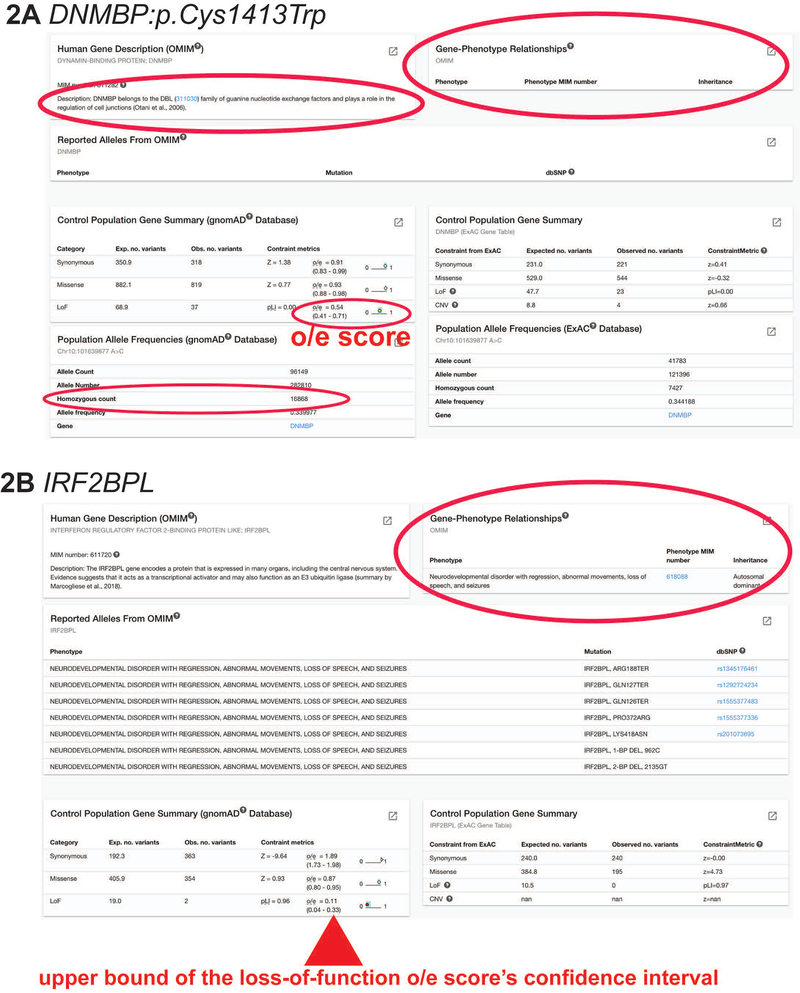
Figure 2: OMIM, gnomAD, and ExAC.
A) Searching for DNMBP:p.Cys1413Trp, MARRVEL’s result page show that there is no known Gene-Phenotype Relationship curated by OMIM, meaning it is not associated with a known disease. Furthermore, this variant is commonly found in the control population database gnomAD. There are 16868 individuals who are homozygous for this variant, indicating that this variant is most likely benign.
B) Searching for IRF2BPL, MARRVEL’s result page show that it is associated with a known autosomal dominant disease called Neurodevelopmental disorder with regression, abnormal movements, loss of speech, and seizures. In the Control Population Gene Summary box for gnomAD database, the pLI score is 0.96, o/e score is 0.11 with the upper bound of 0.33. This means that this gene is highly intolerant of loss-of-function and the loss of one copy of this gene may lead to haploinsufficiency phenotypes.