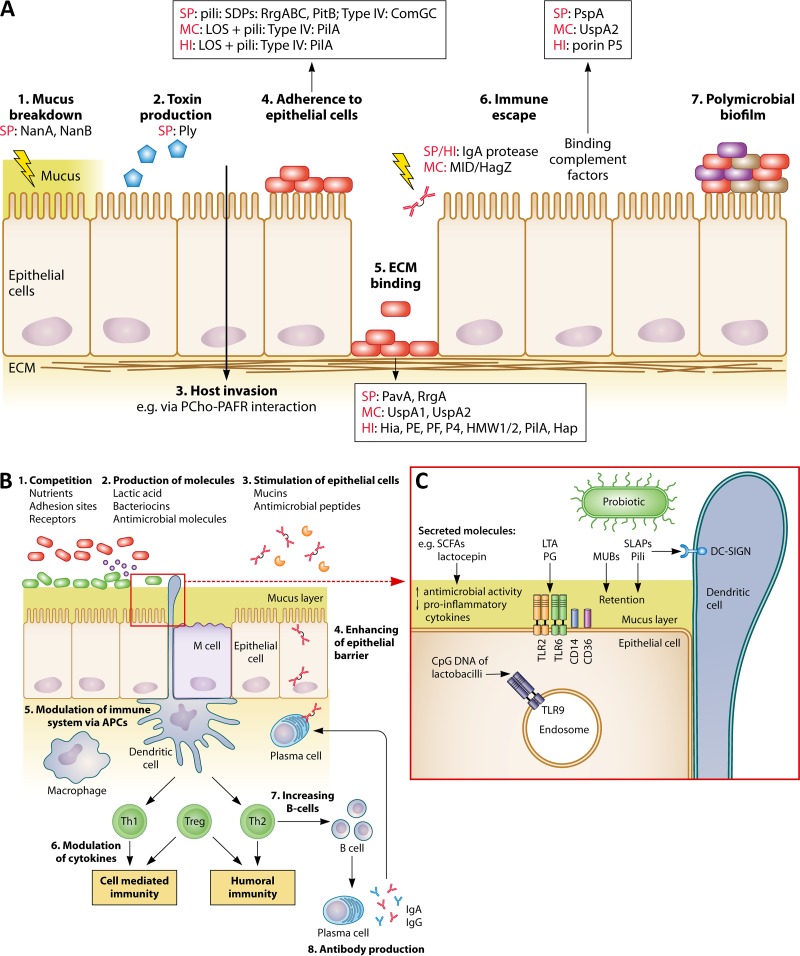
FIG 4.
Comparison between pathogenic and probiotic interactions with the nasopharyngeal epithelium and immune system. (A) Pathogens can interact with nasopharyngeal epithelium and host immune system via (1) breakdown of mucus; (2) production of toxins; (3) invasion of the host; (4) adhesion to the epithelium; (5) binding of ECM via microbial surface components recognizing adhesive matrix molecules (MSCRAMM); (6) escaping immune responses; and (7) the formation of a polymicrobial biofilm. Different frequently occurring pathogenic effector molecules are specified for each (A)OM pathogen. SP, S. pneumoniae; MC, M. catarrhalis; HI, H. influenzae; ECM, extracellular matrix; NanA/B, neuraminidases; Ply, pneumolysin; PCho, phosphorylcholine; PAFR, platelet-activating factor receptor; PavA, pneumococcal adhesion and virulence A; UspA, ubiquitous surface protein; Hia, H. influenzae adhesion; HMW1/2, high-molecular-weight molecules 1/2; Hap, Haemophilus adhesion protein; PspA, pneumococcal surface protein A. It should be noted that not all pathogenic strains or serotypes carry these effector molecules. (B) Postulated beneficial modes of action of URT probiotics. In agreement with beneficial activities in the gut, probiotics could also perform such activities in the URT by, for instance, (1) competition with pathogens for nutrients, adhesion sites, and receptors; (2) production of antimicrobial molecules, such as bacteriocins and lactic acid; (3) stimulation of epithelial cells to modulate mucin and antimicrobial peptide production; (4) enhancement of the epithelial barrier; (5) modulation of the immune system via APCs; (6) modulation of cytokine production; (7) stimulation of increased B-cell production; and (8) stimulation of antibody production. (C) Interaction of several probiotic effector molecules with their receptors localized on the epithelial/dendritic cells or endosomes. APC, antigen-presenting cells; LTA, lipoteichoic acid; MUB, mucus binding protein; PG, peptidoglycan; SCFA, short-chain fatty acids; SLAP, surface-layer-associated protein; Th1/2, T helper 1/2 cells; Treg, regulatory T cells. The figure is based on data described herein in “Infection Mechanisms of the Main Bacterial OM Pathogens” and obtained from references 174, 185, and 281.