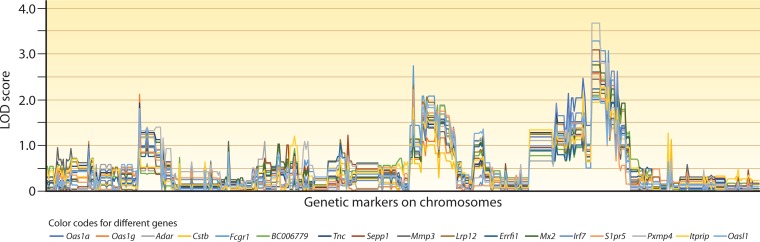
FIG 3.
Plots of host genes based on genome-wide patterns of LOD scores. This figure is based on data published in reference 161. In the study, mice were individually infected with 24 progenies from a genetic cross of Plasmodium yoelii subsp. yoelii 17XNL and Plasmodium yoelii subsp. nigeriensis N67 parasites. mRNA samples from infected spleens were extracted at day 4 postinfection and were hybridized to a microarray representing ∼19,100 unique mouse genes. Transspecies expression quantitative trait locus (ts-eQTL) analysis was used to analyze genome-wide transcription data from the infections against 479 microsatellite (MS) markers typed on the corresponding progenies. This analysis provided a genome-wide pattern of LOD scores (GPLS) for each host gene. Genes with expression levels linked to at least one MS marker (out of 479) with a LOD score of ≥2.0 were chosen, and their GPLSs were clustered based on pattern similarity. This figure shows a group of genes with similar GPLSs including a major peak of LOD score on one end of the parasite chromosome 13. Genes in an individual cluster are often related through roles in the same host response pathways; examples shown in this figure include type I interferon (IFN-I)-stimulated genes (isg) or genes that likely regulate IFN-I responses (Oas1a, Oas1g, Adar, Mx2, Irf7, S1pr5, or Oasl1). The names of the genes in the cluster are indicated under the plot.