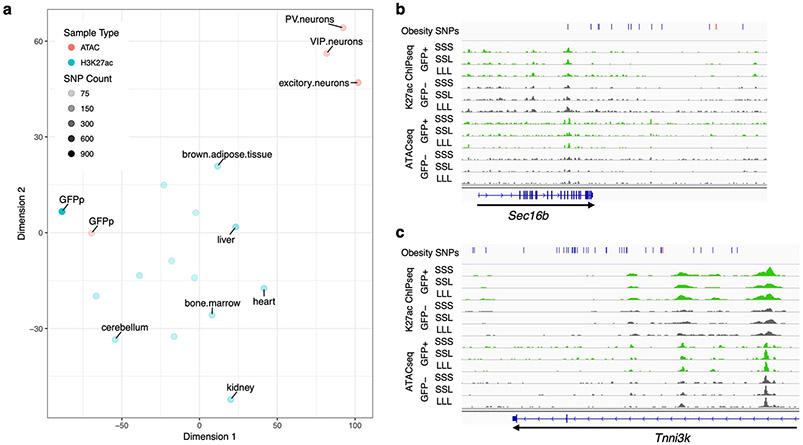
Fig. 6.
Obesity GWAS loci. a tSNE plot showing the differences in obesity-associated SNP-overlaps between tissue and data types. The tSNE plot was generated from a matrix of obesity-associated SNPs which overlap each ChIP-seq or ATAC-seq dataset, such that rows were SNPs and columns samples. Each field was assigned either a 0 (no overlap) or 1 (obesity-associated SNP was overlapped). Color represents the data type: Red – ATAC-seq, Blue – ChIP-seq. The darkness of the color represents the total number of obesity-associated SNPs that overlapped each dataset. b-c Genomic snapshots of Sec16b (b) and Tnni3k (c) loci showing H3K27ac ChIP-seq and ATAC-seq signals in leptin-responsive neurons (green) compared to GFP-negative nuclei (grey). Leptin conditions include mice treated with saline (SSS) or leptin (LLL) injections at 12, 24 and 34 hours (LLL) or saline injections at 12 and 24 hours followed by a single leptin injection at 34 hours (SSL). The obesity SNPs track shows the obesity-associated lead SNP a as red line and in addition SNPs that are in linkage disequilibrium (r2>0.8) as black lines. Each of the two replicates for ChIP-seq and ATAC-seq presented comparable signals as shown.