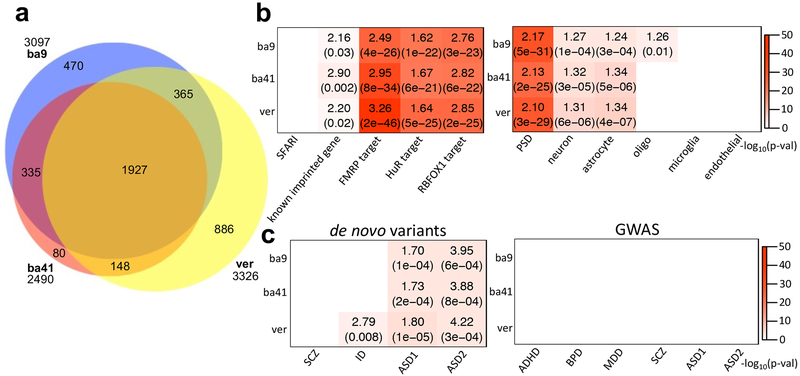
Figure 2.
ASE patterns shared among cases and controls. (a) Venn diagram comparing the overlap of ASE genes from three brain tissues, BA9, BA41, and vermis. (b) Enrichment analyses of the ASE genes with ASD-relevant (SFARI gene list36; Methods) and cell-type specific gene lists (Methods)31. Across all three brain regions, ASE genes showed strong overlap with known imprinted genes (Methods) as well as targets of FMRP28, HuR29, and RBFOX130. Plot showed ORs and FDR corrected p-values for enrichment, if significant. (c) Gene set enrichment study of ASE genes with risk variants in psychiatric disease dataset. From de novo variant datasets45,33, we considered SCZ, intellectual disorder (ID), and ASD. The dataset for SCZ, ID, and ASD1 were gene lists containing de novo likely gene disrupting mutations from the previous study of Iossifov et al.45, and the data for ASD2 represents risk genes integrating de novo copy number variations (FDR≦0.01) from the study of Sanders et al.33 (Methods). The risk variants from GWAS34,35 were considered for ADHD, BPD, MDD, SCZ, and ASD (Methods). Here, ASD1 and ASD2 represent the GWAS datasets from the Cross-Disorder Group of the Psychiatric Genomics34 and Grove et al.35, respectively. If significant, the plot shows ORs and FDR corrected p-values for de novo variant datasets and FDR corrected p-values for GWAS. The sample numbers of BA9, BA41, and vermis are 67, 64, and 64, respectively.