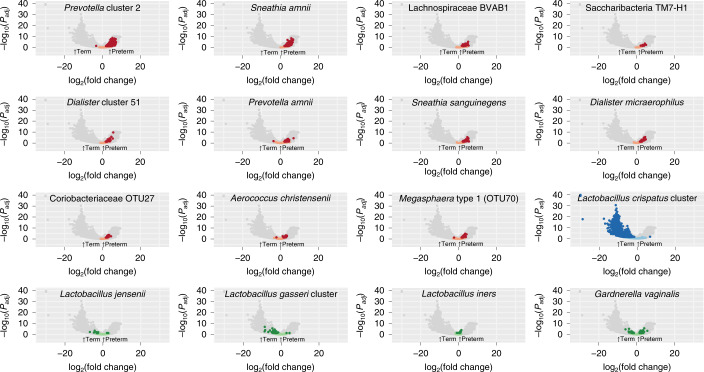
Fig. 4. Volcano plot depicting transcripts that differ greatly between term and preterm cohorts in metatranscriptomics data for taxa of interest.
Analysis was performed by mapping reads from PTB (n = 41) and TB (n = 81) samples to a custom database of genomes representing 56 taxa. Comparative analysis was performed with DESeq2 using a global scaling approach. Genes in candidate PTB taxa identified in 16S rRNA analyses that differ significantly at Padj < 0.05 with a two-sided Wald test, as implemented in DESeq2 with a Benjamini–Hochberg FDR correction, are shown in red and those that were not statistically significant are shown in pink. Genes are also shown for the L. crispatus cluster (associated with TB in the 16S rRNA analyses), with genes that differ significantly (Padj < 0.05) in transcript levels shown in dark blue and those that do not in light blue. Four other common and abundant taxa (L. jensenii, L. gasseri cluster, L. iners and G. vaginalis) are shown, with dark green dots denoting genes that differ significantly (Padj < 0.05) between cohorts and light green dots denoting those that were not statistically significant.