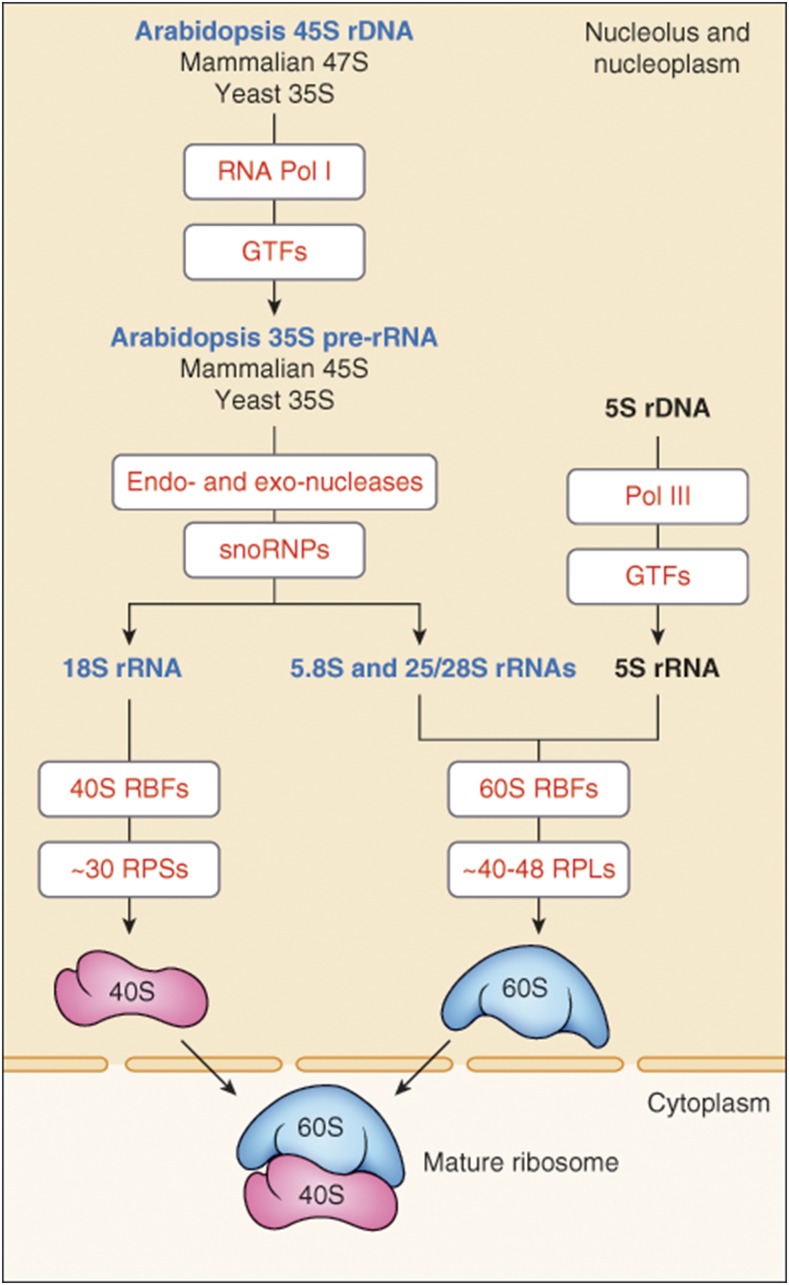
Figure 1.
Graphic Representation of Ribosome Biogenesis in Eukaryotic Cells.
Transcription of rDNA (45S, 47S, and 35S in Arabidopsis, mammals, and yeast, respectively) requires RNA Pol I activity and a subset of GTFs. Transcribed transcript contains the 18S, 5.8S, and 25S (in Arabidopsis and yeast)/28S (in mammals) rRNAs and is first cotranscriptionally processed into pre-rRNA precursor (35S in Arabidopsis and yeast or 45S in mammals). Processing of pre-rRNAs into mature 18S, 5.8S, and 25S/28S rRNA involves multiple endonucleolytic and exonucleolytic cleavages and the modification of numerous rRNA residues, mainly pseudouridylation and 2′-O-ribose methylation, by snoRNP complexes (see Figure 3). 18S rRNAs assemble with ribosomal proteins RPSs (S for small) to form 40S ribosome subunits, while 5.8S and 25S/28S assemble with ribosomal proteins RPLs (L for large) and 5S rRNA transcribed by RNA Pol III. Assembly and transport of ribosomal particles from the nucleolus to the cytoplasm requires hundreds of specific 40S and 60S RBFs (Figures 4 and 5; Supplemental Data Sets 3 and 4). The 40S and 60S ribosomal subunits join to form translationally competent ribosomes.