Figure 7.
Identification of the GmSIN1 Binding Motif (SIN1BM).
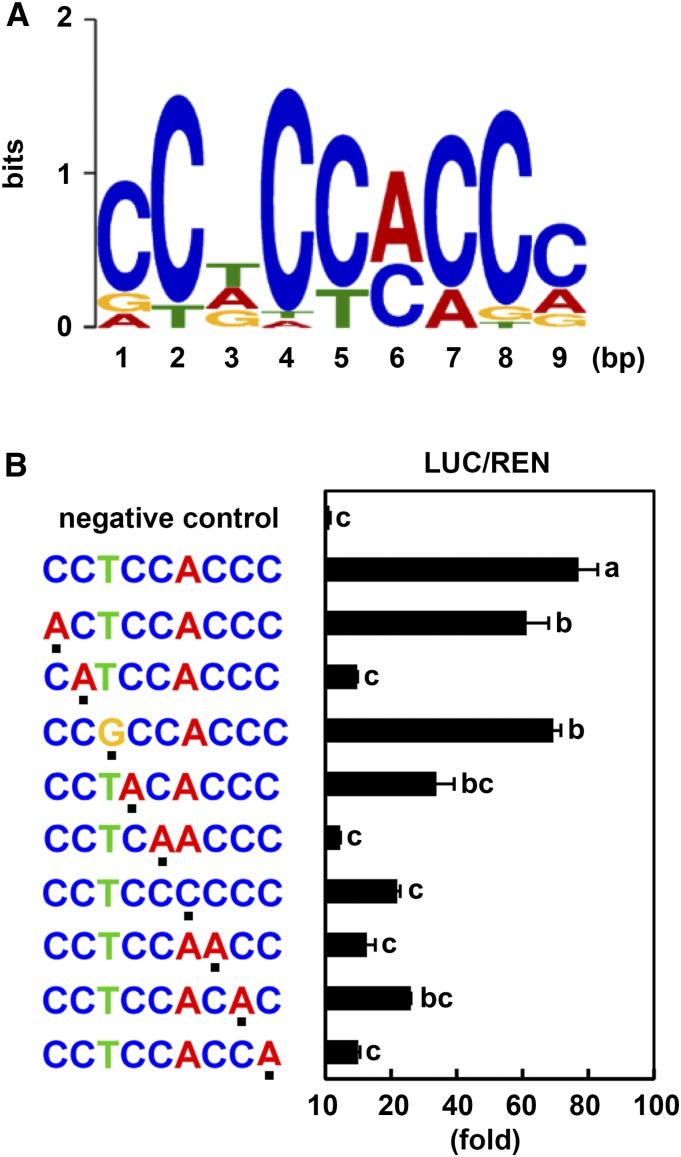
(A) Using MEME analysis, conserved sequences were identified in the promoter regions of genes co-upregulated in GmSIN1 OE plants and the NaCl-treated wild-type plants (Table 2). Position weight matrix of SIN1BM showing the probability of a nucleotide(s) at each position.
(B) Interaction of GmSIN1 with the SIN1BM consensus and its substituted sequences in LUC (Firefly luciferase) assays. The sequences of interest were fused to a promoter fragment of TUB2 that does not contain a SIN1BM. LUC activity and REN (Renilla luciferase) activity were measured. Dots below the sequences indicate the substitution from SIN1BM, which was determined by MEME analysis (shown in [A]). pTUB2pro::LUC and pGreenII 0800-35S-LUC were used for negative control and positive control, respectively. Mean and se of LUC activity/REN activity were obtained from more than four independent transformation experiments. Significant differences between samples labeled with different letters (a, b, c) were determined by one-way ANOVA and Tukey’s test, P < 0.05.