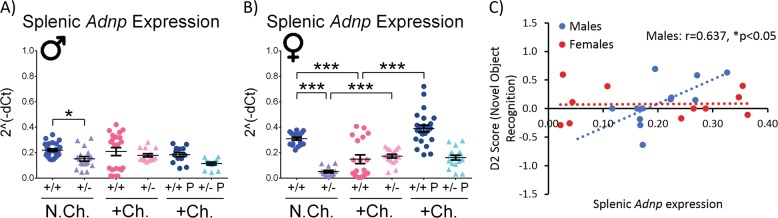
Fig. 3. Splenic Adnp gene expression is regulated in a sex-dependent manner, as well as by PACAP treatment in females, with cognition positively correlating splenic Adnp expression.
Two-way ANOVA with Tukey post hoc test was performed (males (M): N.Ch. Adnp+/+ N = 12–26; N.Ch. Adnp+/− N = 12–18; +Ch. Adnp+/+ N = 21; +Ch. Adnp+/− N = 15; +Ch. Adnp+/+ PACAP N = 14; +Ch. Adnp+/− PACAP N = 12; females (F): N.Ch. Adnp+/+ N = 18; N.Ch. Adnp+/− N = 12; +Ch. Adnp+/+ N = 19; +Ch. Adnp+/− N = 17; +Ch. Adnp+/+ PACAP N = 24; +Ch. Adnp+/− PACAP N = 17). For gene expression analysis, results are presented as 2−ΔCT, normalized to Hprt. For correlation testing, if both plotted data sets were normally distributed, a Pearson’s correlation analysis was performed. If at least one of the data sets was not normally distributed, a Spearman’s correlation was performed. a Splenic Adnp expression in vehicle-treated males, significant main genotype effect (F(1,76) = 5.664, p = 0.020) was found, with significant differences between non-challenged Adnp+/+ and Adnp+/− mice (*p < 0.05). b Splenic Adnp expression in stressed-challenged females: main genotype (F(1,73) = 15.336, p < 0.001), treatment (F(1,73) = 19.029, p < 0.001), and interaction (F(1,73) = 23.179, p < 0.001) effects were found, with significant differences between challenged Adnp+/+ and Adnp+/− mice (***p < 0.001), as well as challenged PACAP- vs. vehicle-treated Adnp+/+ mice (***p < 0.001). In vehicle-treated females, significant main genotype (F(1,62) = 26.464, p < 0.001) and interaction (F(1,62) = 38.150, p < 0.001) effects was found, with significant differences between non-challenged Adnp+/+ and Adnp+/− mice (***p < 0.001), challenged vs. non-challenged Adnp+/− mice (***p < 0.001), and challenged vs. non-challenged Adnp+/+ mice (***p < 0.001). c In non-challenged males (N = 12), splenic Adnp gene expression was positively correlated with the D2 score in the novel object recognition test (Pearson’s correlation, r = 0.637, *p < 0.05)