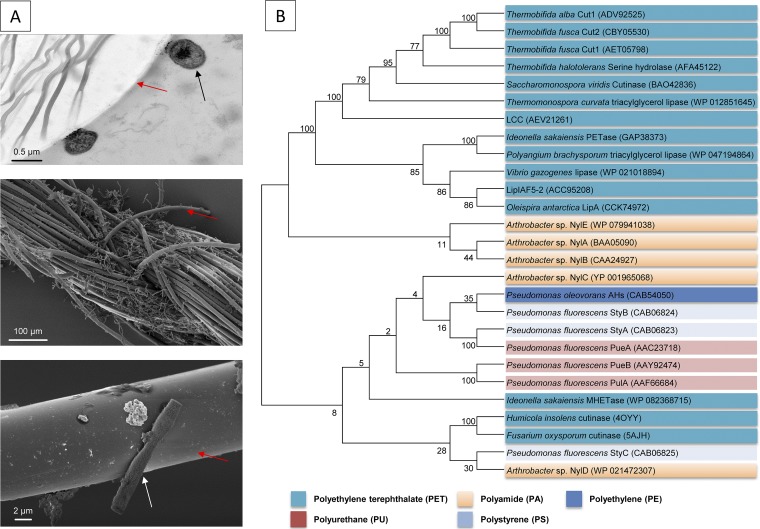
FIG 2.
(A) Electron microscopic images of Comamonas sp. strain DDHH 01 attached and hydrolyzing PET fibers. Comamonas sp. DDHH 01 was isolated from a sewage enrichment culture. Red arrows indicate PET fibers. Black and white arrows indicate bacterial cells. (Top) Transmission electron microscopy image of a PET fiber with attached Comamonas sp. cells. (Middle) Scanning electron microscope image of PET yarn with microcolonies. (Bottom) Closeup of a single cell on the surface of a single PET fiber. (B) Topology of a neighbor-joining tree containing representative sequences of most of the currently known synthetic polymer- or oligomer/monomer-degrading enzymes. The tree is based on amino acid sequence homologies. Overall, 27 known functional and verified enzymes were included in this alignment. This represents the majority of the currently known and biochemically characterized enzymes. PET hydrolases represent the largest fraction of known and studied enzymes. The alignment was calculated using T-Coffee in accurate mode (124). The tree was calculated with Molecular Evolutionary Genetics Analysis version 6 (MEGA6) (125) and is not rooted. A similarity and identity matrix for all included sequences, together with their accession numbers, is provided in the supplemental material (Table S1).