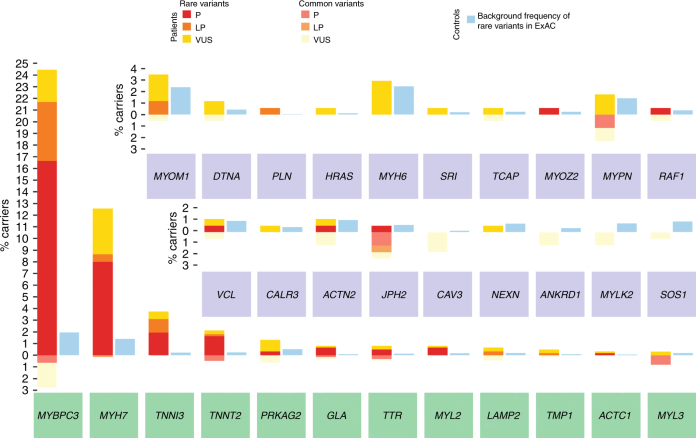
Fig. 2. Cumulative variant frequencies observed in hypertrophic cardiomyopathy patients compared with those in the Exome Aggregation Consortium.
Proportion of hypertrophic cardiomyopathy (HCM) probands evaluated by next-generation sequencing (n = 613) carrying clinically reportable variants (left-hand side bar for each gene). Proportions are divided into “rare” (minor allele frequency (MAF) < 0.01% in the Exome Aggregation Consortium (ExAC); above 0 on the y axis) and “common” (MAF > 0.01% in ExAC; below 0 on the y axis). Variants with MAF > 0.01% are unlikely to be truly disease causing, as recently demonstrated.26 Variation in HCM is also divided into pathogenic (P: red, rare; pale red, common), likely pathogenic (LP: orange, rare; pale orange, common), and variants of uncertain significance (VUS: yellow, rare; pale yellow, common). Light blue bars (right-hand side for each gene) represent the proportion of rare protein-altering variant carriers in ExAC. Robustly validated disease genes (turquoise background) were sequenced in all probands screened using next-generation sequencing (n = 613), while other genes (purple background) were sequenced on a subset of 171 probands (Fig. 1). Genes associated with syndromic forms of HCM are indicated in bold. For example, MYBPC3 is characterized by features typical of a major disease gene: strong rare variation excess in patients (left-hand side bar) versus ExAC (right-hand side bar), reflective of high diagnostic interpretability, and a high proportion of P/LP variants (red/orange), indicating high actionability for cascade screening (see Materials and Methods for derivation). Conversely, MYPN is characterized by all variants originally classified as P/LP being too common in ExAC to be pathogenic, the frequency of rare variant carrier patients being essentially equal to the background population level (low diagnostic interpretability), and all rare variants in patients being VUS (null actionability for cascade screening)