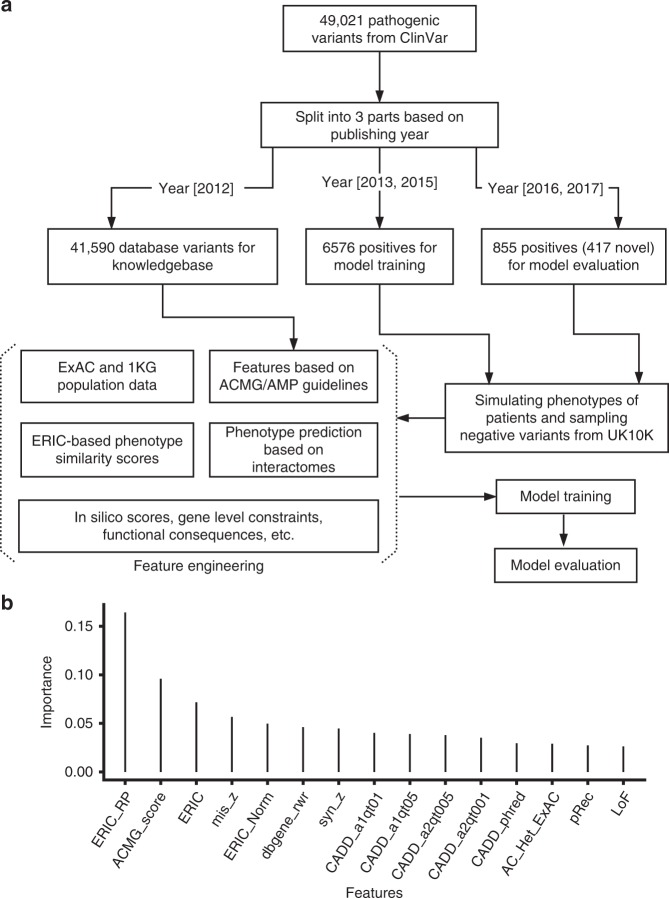
Fig. 1.
Schematic overview of the Xrare model and most important features for predicting variant pathogenicity. a Schematic overview of the Xrare model. The collected 49,021 pathogenic ClinVar variants were divided into three parts in terms of their publishing years: 41,590 variants identified by 2011 used for implementing American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) guideline evidence; 6576 variants spiked in synthetic genomes for model training; the remaining 855 variants identified since 2016 used for model evaluation. The preexisting in silico computation scores of variants, population-level scores, and other ACMG evidence scores were used as features for machine learning. Phenotype-related features came from gene–phenotype similarity Emission-Reception Information Content (ERIC) score and predicted gene–phenotype associations. b The top 15 most important features from Xrare model. See Supplementary Figure S1 for all features.