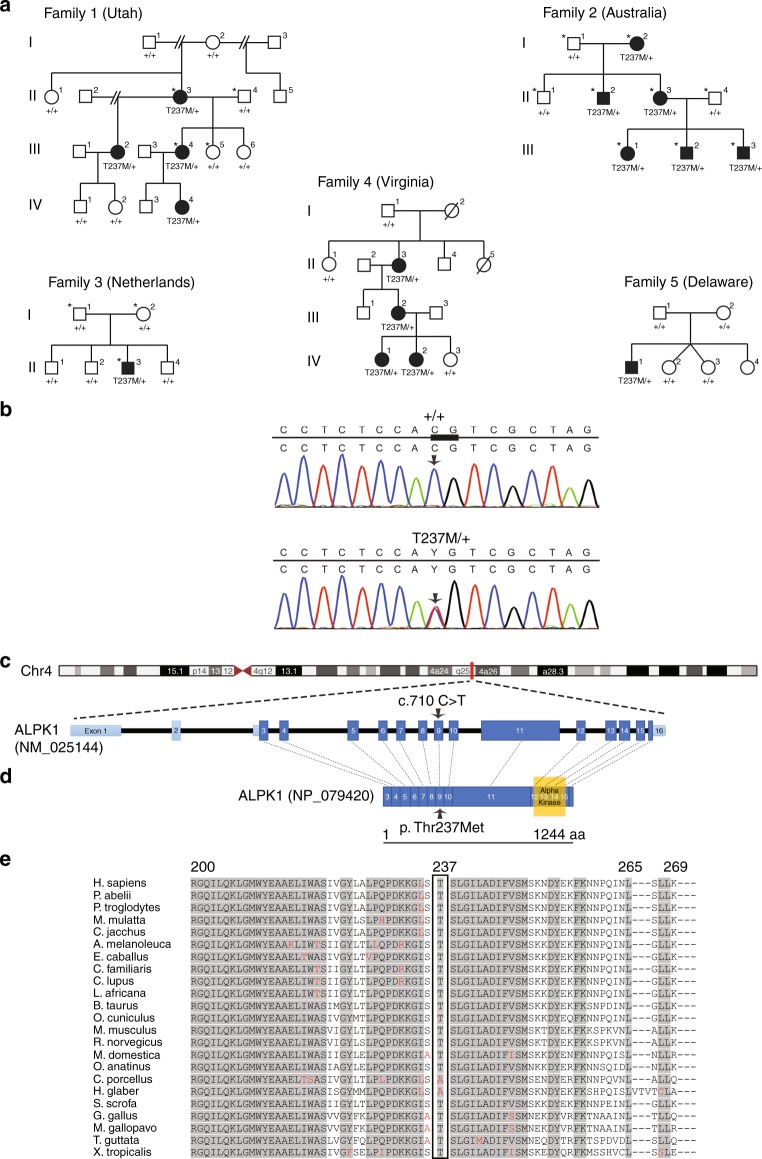
Fig. 1.
Pedigree of families with ROSAH syndrome and pathogenic variant identification in ALPK1. (a) Five families affected by autosomal dominant ROSAH syndrome were analyzed in this study, from Utah (USA), Australia, The Netherlands, Virginia (USA), and Delaware (USA). Affected individuals are shaded and those examined by exome sequencing and genome sequencing in the Utah, Australia, and Netherlands families are indicated with an asterisk. Variant status is indicated below each individual for whom sequencing data was obtained (T237M indicates allele coding for [p.Thr237Met]; + indicates normal allele). (b) Subsequent Sanger sequencing in all families confirmed segregation of the ALPK1 (NM_025144) variant with disease. All individuals with the variant in ALPK1 have the ROSAH syndrome phenotype. The c.710C base pair (bp) is indicated by the arrow, and the bp 710–711 CG dimer is indicated by the thickened line. (c) Exonic structure of ALPK1 (NM_025144) with the pathogenic variant on exon 9 identified in ROSAH syndrome. (d) Protein structure of ɑ-kinase 1 showing the location of the pathogenic variant and the location of the known kinase region. (e) Comparison across species for ALPK1 Thr237 and flanking sequences using multiple sequence alignment from UniProtKB/UniRef100 Release 2011_12 (14 December 2011) database. A high degree of conservation of the Thr237 amino acid in vertebrates is evident. Flanking sequences are also highly conserved. Shaded areas represent consensus sequence. Red letters represent deviation from consensus.