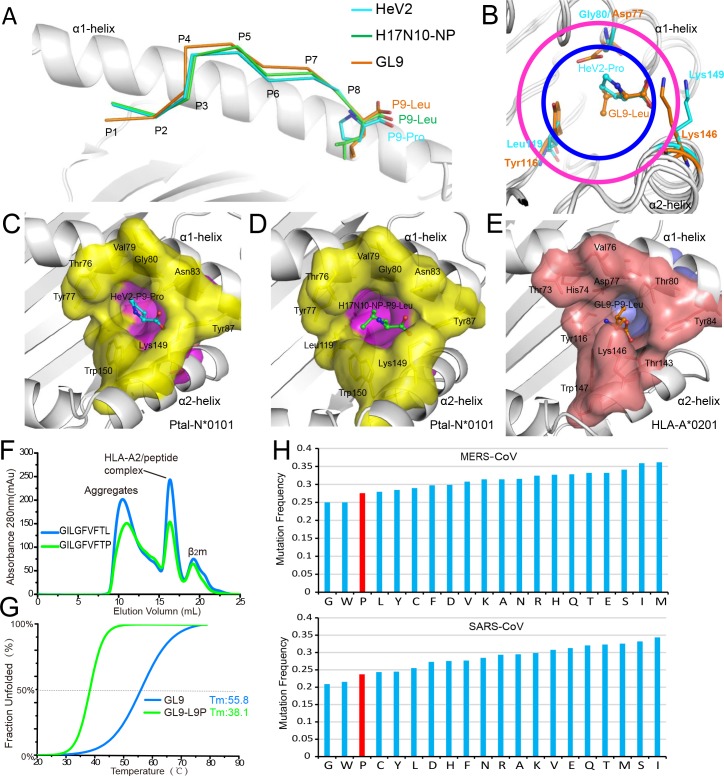
Fig 6. The extraordinary preference for Pro as the PΩ anchor of Ptal-N*01:01–binding peptides.
(A) The superimposition of 9-mer peptides HeV2 (cyan) and H17N10-NP (green) presented by Ptal-N*01:01 and GL9 (orange) presented by HLA-A*0201 (PDB code: 3I6G). The heavy chains are represented as gray cartoons. The PΩ anchors of peptides are shown as sticks, and the side chains of other residues of the peptides are omitted. (B) Distinct F pockets of Ptal-N*01:01 and HLA-A*0201. The heavy chains are represented as gray loops. Gly80, Leu119, and Lys149 of Ptal-N*01:01 are represented as cyan sticks. The corresponding residues of HLA-A*0201 are shown as orange sticks. The given entrances for F pockets of Ptal-N*01:01 and HLA-A*0201 are labeled by purple and deep blue circles, respectively. (C-D) The side chains of PΩ-Pro from peptide HeV2 (C) and PΩ-Leu from peptide H17N10-NP (D) insert into the F pockets of Ptal-N*01:01. The F pocket of Ptal-N*01:01 is indicated in surface representation with a yellow entrance and purple bottom. (E) The side chain of residue PΩ-Leu from peptide GL9 inserts into the F pocket of HLA-A*0201. The F pocket of HLA-A*0201 is indicated by the representation of a brown entrance with a light blue bottom. (F-G) The stabilities of HLA-A*0201 complexed with peptide GL9 and its mutant GL9-L9P (with a Pro at the P9 position) were evaluated by in vitro refolding (F) and CD spectroscopy (G). The numerical data are included in S1 Data. (H) The relatively lower mutation frequency of amino acid Pro (red bar) as Try and Gly among the 20 component amino acids in the proteins of MERS-CoV and SARS-CoV. The mutation frequency = the number of overall mutations for each amino acid/(the number of occurrences of the amino acid in the reference sequence×total number of sequences). The proteomes of 1,000 MERS-CoV genomes and SARS-CoV genomes were retrieved from GenBank, respectively. The detailed information is included in S2 Data. CD, circular dichroism; GL9, GILGFVFTL; HeV, Hendra virus; MERS-CoV, Middle East respiratory syndrome coronavirus; PDB, Protein Data Bank; SARS-CoV, severe acute respiratory syndrome coronavirus; Tm, midpoint transition temperature.