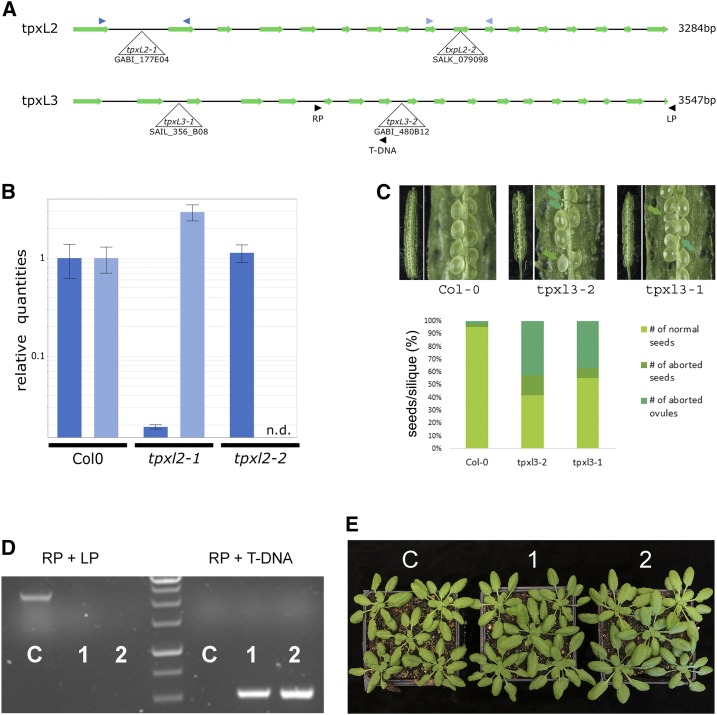
Figure 5.
Mutant analysis revealed TPXL2 as nonessential whereas TPXL3 is essential for embryo development. A, Schematic overview of the gene models for TPXL2 (top) and TPXL3 (bottom) with indication of the positions of the T-DNA insertion alleles analyzed. Primers used for RT-qPCR and genotyping analysis are indicated by color-coded arrowhead pairs. B, RT-qPCR analysis of homozygous tpxl2 mutants showing the absence of full-length transcripts over the T-DNA positions (n.d., not detected). Error bars = se. C, Representative silique pictures and quantification of seed development of selfed tpxl3-1 (+/−) and tpxl3-2 (+/−) plants grown together with control plants (Col-0). Both TPXL3 alleles show a high percentage of aborted ovules and some aborted seeds. The quantification shows the combined result of different siliques from different plants. For both Col-0 and tpxl3-2, 20 siliques were analyzed (from two different plants each), whereas for tpxl3-1, we analyzed 30 siliques (from three different plants). D and E, Introducing TPXL3-GFP into tpxl3-2(+/−) mutants allowed identification of homozygous tpxl3-2 mutants (two independent lines are shown; “1” and “2”), which develop similarly to the wild-type controls (“C”). Genotyping results revealed the absence of wild-type (LP + RP) fragment (∼2.5-kb) and the presence of the T-DNA specific fragment (RP + T-DNA; ∼0.5-kb) in two independent lines carrying the TPXL3-GFP transgene (“1” and “2”) in contrast to control plants (“C”).