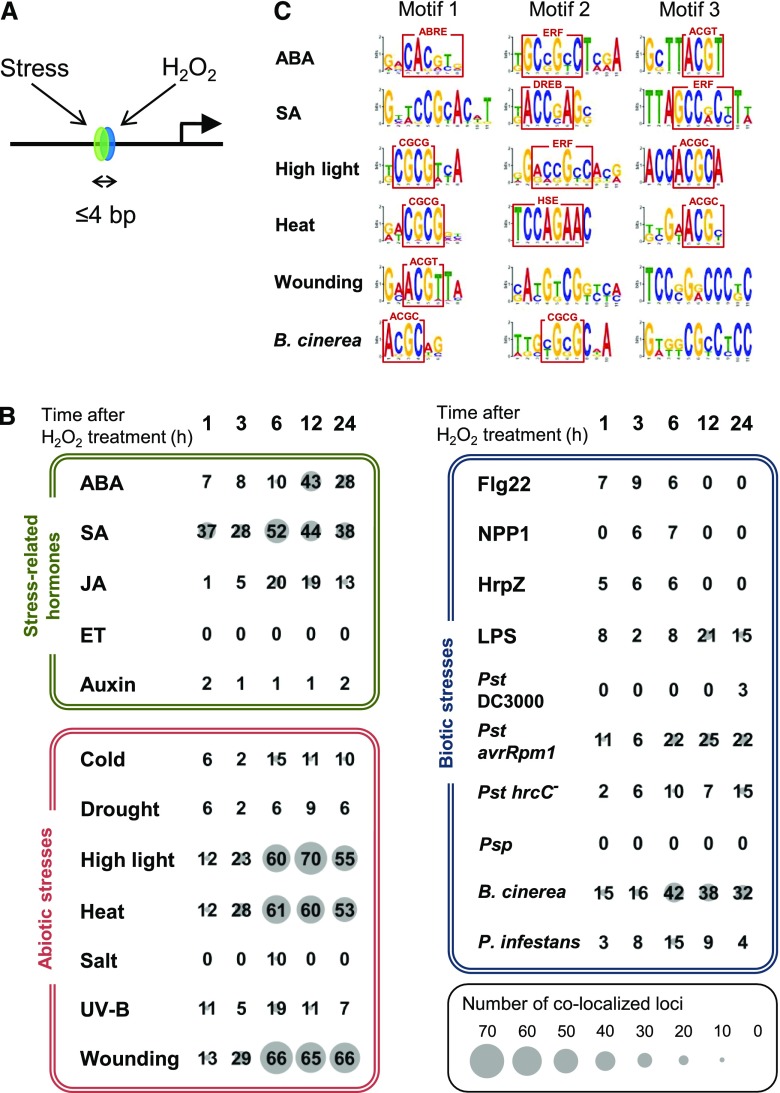
Figure 3.
Colocalization analysis of predicted cis-elements for H2O2 and other responses. A, Putative cis-elements identified by two independent predictions colocalizing within 4 bp (>3 bp overlap) were counted. B, The numbers in matrices indicate colocalized loci for two independent cis predictions. One prediction is for the H2O2 response, and the induction time after H2O2 treatment (h) used for the prediction is shown at the top (1–24). The second prediction includes three categories, stress-related hormone responses (green box), abiotic stress responses (red box), and biotic stress responses (blue box). The numbers at the cross point between the first horizontal prediction and the second vertical prediction represent loci of the corresponding crosstalk. The size of the gray circle behind a number gives a visual representation of the degree of colocalization. Time point of microarray data for promoter prediction was selected. ABA, 1 h; SA, 3 h; JA (treated with methyl jasmonate), 3 h; ET (treated with the metabolic precursor, 1-aminocyclopropane-1-carboxylic acid), 3 h; Auxin (treated with IAA), 3 h; Cold, 24 h; Drought, 1 h; High light, 3 h; Heat, 3 h; Salt, 3 h; UV-B, 3 h; Wounding, 3 h; flagellin22 (Flg22), 1 h; necrosis-inducing Phytophthora protein (NPP1), 1 h; harpinZ (HrpZ), 1 h; LPS (lipopolysaccharide), 4 h; Pst DC3000 (Pseudomonas syringae pv. tomato DC3000, 6 h; Pst avrRpm1 (P. syringae pv. tomato carrying an aviruent gene), 6 h; Pst hrcC− (P. syringae pv. tomato hrcC mutant), 24 h; Psp (P. syringae pv. phaseolicola), 6 h; B. cinerea, 18 h; P. infestans, 24 h. C, Sequences at the colocalization of double predictions were mixed for each second prediction, and subjected to motif extraction by Multiple Em for Motif Elicitation (http://meme-suite.org/tools/meme). Motifs in the red boxes are ABRE (Hattori et al., 2002), ERF (Hao et al., 1998), DREB (Sakuma et al., 2002), CGCG box (Yang and Poovaiah, 2002), HSE (Barros et al., 1992), core sequence recognized by bZIP family proteins (ACGT; Foster et al., 1994), and ACGC, which is a novel motif. See Supplemental Table S12 for accession numbers of microarray data using promoter prediction.