TPK3 is a tonoplast K+ channel that does not function in photosynthesis, unlike the bona fide thylakoid K+/H+ exchanger KEA3 and the thylakoid Cl− channel VCCN1.
Abstract
Photosynthesis is limited by the slow relaxation of nonphotochemical quenching, which primarily dissipates excess absorbed light energy as heat. Because the heat dissipation process is proportional to light-driven thylakoid lumen acidification, manipulating thylakoid ion and proton flux via transport proteins could improve photosynthesis. However, an important aspect of the current understanding of the thylakoid ion transportome is inaccurate. Using fluorescent protein fusions, we show that the Arabidopsis (Arabidopsis thaliana) two-pore K+ channel TPK3, which had been reported to mediate thylakoid K+ flux, localizes to the tonoplast, not the thylakoid. The localization of TPK3 outside of the thylakoids is further supported by the absence of TPK3 in isolated thylakoids as well as the inability of isolated chloroplasts to import TPK3 protein. In line with the subcellular localization of TPK3 in the vacuole, we observed that photosynthesis in the Arabidopsis null mutant tpk3-1, which carries a transfer DNA insertion in the first exon, remains unaffected. To gain a comprehensive understanding of how thylakoid ion flux impacts photosynthetic efficiency under dynamic growth light regimes, we performed long-term photosynthesis imaging of established and newly isolated transthylakoid K+- and Cl−-flux mutants. Our results underpin the importance of the thylakoid ion transport proteins potassium cation efflux antiporter KEA3 and voltage-dependent chloride channel VCCN1 and suggest that the activity of yet unknown K+ channel(s), but not TPK3, is critical for optimal photosynthesis in dynamic light environments.
Photosynthesis provides metabolic energy for nearly all life on earth. During this process, light is utilized for CO2 fixation, growth and additional energy-dependent metabolic pathways. In oxygenic photosynthesis, light energy induces charge separations at two photosystems. At PSII, electrons are stripped from water molecules releasing protons into the lumen. These electrons move along the thylakoid electron transport chain toward PSI and finally reduce NADP+ to NADPH. During electron transport, protons are translocated from the chloroplast stroma into the thylakoid lumen, generating a proton motive force (pmf) which drives the ATP synthase to produce ATP. The thylakoid pmf is comprised of two components, a pH gradient (ΔpH) and a membrane potential (Δψ). In addition to providing the driving force for ATP synthesis, the luminal proton concentration has important regulatory functions. Above a certain proton concentration threshold, a photoprotective mechanism is activated that dissipates excess absorbed light energy as heat. This mechanism is called energy-dependent quenching (qE). qE involves the PsbS protein and the xanthophyll cycle and is the largest and fastest contributor to nonphotochemical quenching (NPQ) of chlorophyll a (Chl a) fluorescence (Jahns and Holzwarth, 2012).
In nature, availability of light energy to drive photosynthesis undergoes frequent and rapid changes. Upon transition to high-light intensities, rapid activation of qE and photosynthetic control allows plants to adjust photosynthesis and mitigate damage to the photosystems (discussed in Murchie and Niyogi, 2011). The rapid deactivation of qE maximizes photochemistry during the transition to light-limiting conditions. Accelerating the dynamics of NPQ has been hypothesized as a potential mechanism to improve photosynthetic efficiency and biomass production in plants (Zhu et al., 2004). Indeed, transgenic tobacco (Nicotiana tabacum) plants with increased PsbS level and an accelerated xanthophyll cycle possessed faster NPQ kinetics and yielded up to 15% more biomass in the field (Kromdijk et al., 2016). An alternative way to adjust NPQ dynamics is to modify thylakoid ion transport and thus pmf composition. During a transition to low light, the K+/H+ antiporter KEA3 shifts the composition of pmf toward higher Δψ, thereby deactivating NPQ (Armbruster et al., 2014). Conversely, during a transition from low to high light, the dissipation of Δψ by ion channels leads to a larger fraction of pmf being stored as ΔpH (i.e. ΔpH increases), which in turn is required to activate the NPQ process (Davis et al., 2016). Lack of the thylakoid Cl− channel VCCN1/Best1 has been shown to increase levels of Δψ, thereby decreasing levels of NPQ upon sudden high light exposure, whereas VCCN1 overexpression results in lower Δψ and higher NPQ (Duan et al., 2016; Herdean et al., 2016). Moreover, it was shown that a high Δψ across the thylakoid membrane leads to PSII photo-inhibition (qI; the slow-relaxing component of NPQ) by favoring charge recombination (Davis et al., 2016). Thus, thylakoid ion channels may additionally be important to avoid PSII photoinhibition.
Another ion channel that has been described as a thylakoid membrane protein is TPK3 (Zanetti et al., 2010), one of the five members of the tandem-pore K+ channel family in Arabidopsis (Arabidopsis thaliana; Hedrich, 2012). Since initially no transfer DNA (T-DNA) insertion lines in the TPK3 locus were available (Checchetto et al., 2012), tpk3 RNA interference (RNAi) lines were analyzed (Carraretto et al., 2013). Specifically, under moderate growth light conditions of 90 or 100 μmol photons m−2 s−1, tpk3 RNAi plants showed an increased fraction of pmf stored as Δψ and less pronounced NPQ induction resembling the vccn1 loss-of-function mutants (Carraretto et al., 2013; Allorent et al., 2018). So far, photosynthesis in TPK3-deficient or gain-of-function mutants has not been analyzed during reoccurring sudden increases in light intensity (i.e. low- to high-light transitions). A thylakoid K+ channel-deficient mutant is expected to phenotypically resemble the thylakoid chloride channel-deficient mutant vccn1, as both luminal Cl− influx and K+ efflux have been shown to occur in response to light exposure (Hind et al., 1974). Here, we obtained a tpk3 T-DNA insertion line to analyze the effects of TPK3 loss of function on photosynthesis under dynamic light environments.
RESULTS
TPK3 Fluorescent Protein Fusions Localize to the Tonoplast of Nicotiana benthamiana and Arabidopsis Cells
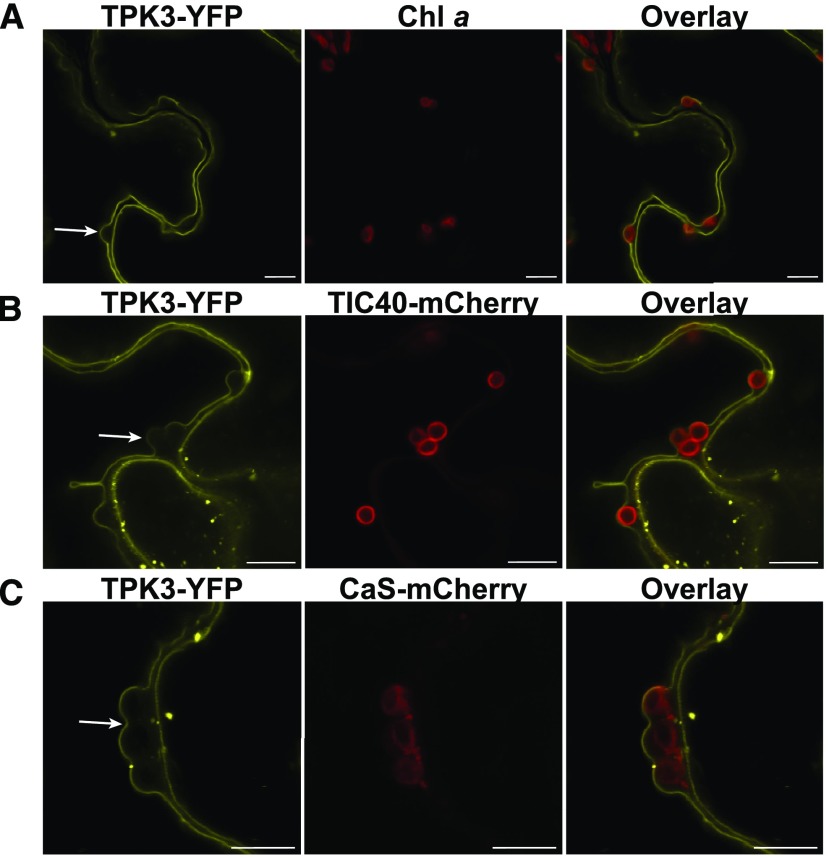
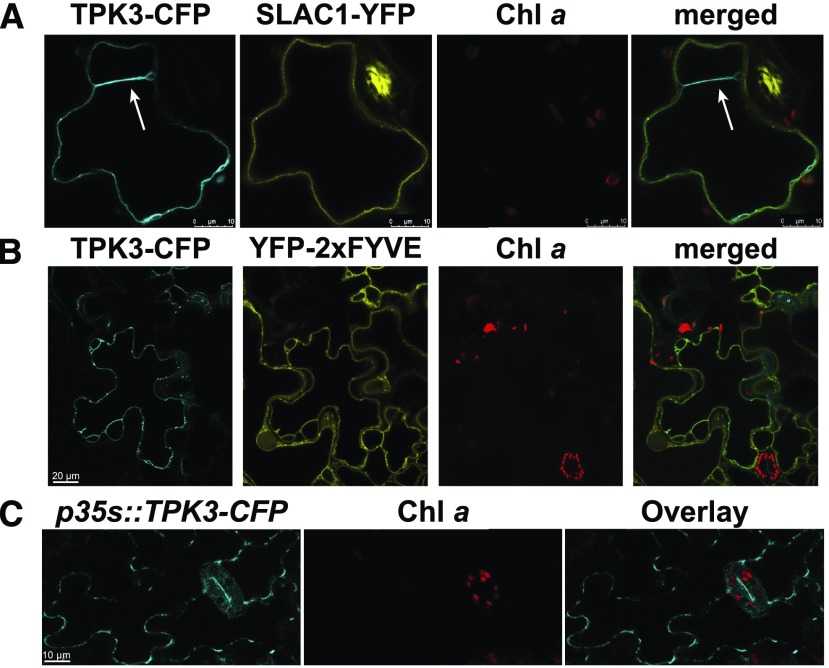
Over the last years, contradictory results on the subcellular localization of TPK3 channel fluorescent fusion proteins have been reported that suggest localization to either the tonoplast (Voelker et al., 2006; Dunkel et al., 2008; Maîtrejean et al., 2011) or the chloroplast thylakoid membrane (Carraretto et al., 2013). To determine TPK3 localization in an independent approach, we expressed two TPK3 fluorescent protein fusions (i.e. TPK3-yellow fluorescent protein [YFP] and TPK3-cyan fluorescent protein [CFP]) transiently in N. benthamiana leaf cells. In both cases, the TPK3 signal was observed distinctly apart from the Chl a fluorescence (Figs. 1 and 2). The fluorescence signal of the TPK3 fusions originated exclusively from a membrane outside the chloroplast (Fig. 1A). This finding was further corroborated by colocalization with the well-established chloroplast markers AtTIC40-mCherry, marking the inner envelope (Fig. 1B; Supplemental Movie S1), and CaS-mCherry, which contains the thylakoid membrane-localized Ca2+ sensor protein (Nomura et al., 2008; Vainonen et al., 2008; Weinl et al., 2008; Fig. 1C). Again, the TPK3-YFP signal was absent from the chloroplast and did not overlay with either the envelope or thylakoid membrane markers (Supplemental Fig. S1A). In fact, the membrane that contained the TPK3 signal appeared to press the chloroplast against the cell boundary, which suggests a localization of TPK3 in the central vacuolar membrane. To determine the exact membrane localization of TPK3, we carried out further colocalizations using TPK3-CFP either in combination with the plasma membrane Slow-Anion Channel 1 (SLAC1; mVenus-SLAC1-YFP; Vahisalu et al., 2008; Fig. 2A) or the tonoplast marker YFP-2xFYVE (Fab1, YOTB, Vac1, and EEA1 domain-containing protein; Singh et al., 2014; Fig. 2B). From these studies, it became apparent that the TPK3 fluorescence signal aligned perfectly with the tonoplast marker FYVE but not with plasma membrane channel SLAC1 (Fig. 2A, arrow).
Figure 1.
TPK3 fluorescent fusion proteins are localized outside of the chloroplast. A, TPK3-YFP and Chl a fluorescence are shown. Clearly, the TPK3-YFP signal remained outside the chloroplasts. Scale bar = 20 µm. B and C, TPK3-YFP localization analysis alongside bona fide chloroplast controls TIC40-mCherry (B; inner envelope) and CaS-mCherry (C; thylakoid membrane) confirm that TPK3 does not localize to the chloroplasts. For all localization studies, constructs were transiently expressed in N. benthamiana leaves.
Figure 2.
TPK3 colocalizes with a tonoplast marker. A and B, TPK3-CFP does not colocalize with the plasma membrane control SLAC1-YFP signal (A, arrow) but rather with the tonoplast marker YFP-2xFYVE (B). Scale bar = 20 μm. C, In stable Arabidopsis p35s::TPK3-CFP overexpressor lines, TPK3-CFP fluorescence signal never originated from leaf chloroplast, but rather from the tonoplast. Chl a fluorescence is shown in red. Scale bar = 10 µm.
To verify TPK3 protein localization in Arabidopsis, we isolated stable Cauliflower mosaic virus 35S promoter (p35s)::TPK3-CFP overexpression lines in the Columbia (Col-0) wild type (Supplemental Fig. S1, B and C). The TPK3 localization confirmed our observations from transiently expressing N. benthamiana cells, i.e. the TPK3-CFP signal was only detected in the vacuolar membrane and never aligned with the thylakoid Chl a fluorescence (Fig. 2C).
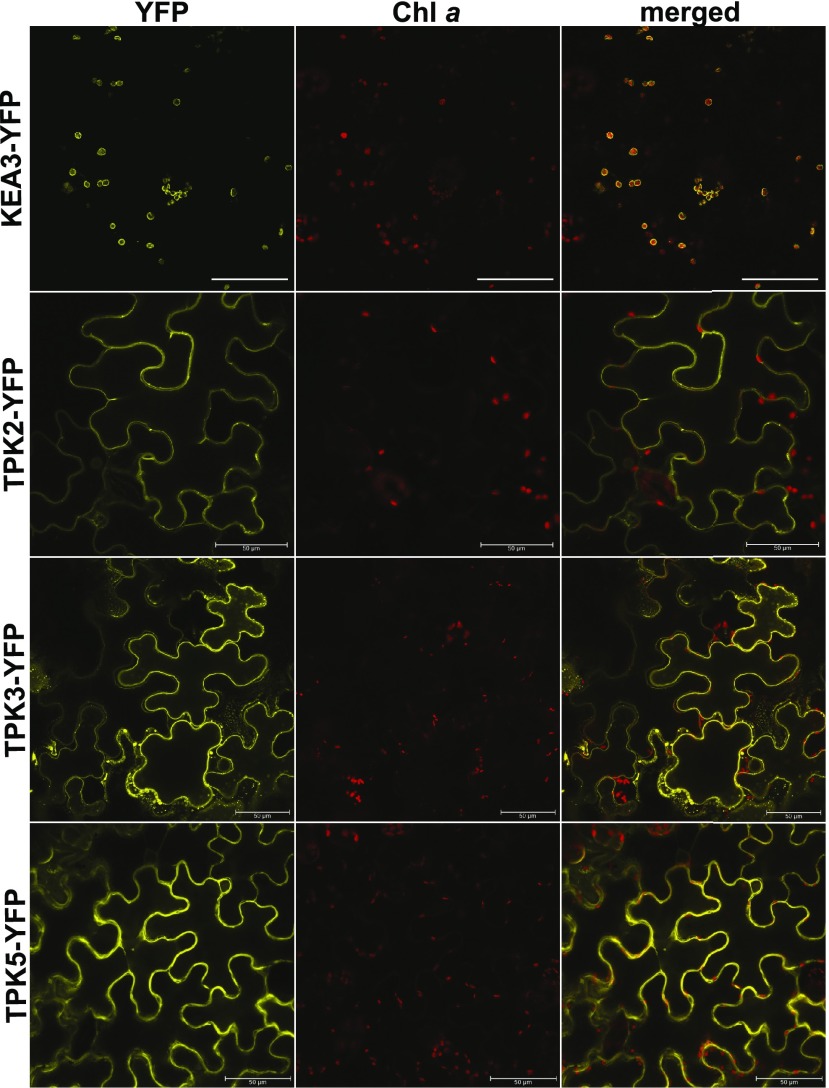
The tandem-pore K+ cation channel family consists of five members in Arabidopsis. According to the phylogenetic relationship among TPK family members, the two members TPK1 and TPK4 form their own subclades, whereas TPK2, TPK3, and TPK5 cluster together into one subclade (Hedrich, 2012; Supplemental Fig. S2). The TPK2-YFP and TPK5-YFP fusion proteins showed a localization pattern highly similar to that of TPK3, suggesting that all members of this subclade have a function in the tonoplast, as reported initially (Voelker et al., 2006; Dunkel et al., 2008; Fig. 3). None of three TPK members resembled the signal observed for thylakoid marker KEA3-YFP in N. benthamiana (Fig. 3).
Figure 3.
TPK2 and TPK5, the closest TPK3 homologs, are also localized at the tonoplast. Constructs were transiently expressed in N. benthamiana leaves. Signals observed in TPK2-YFP-, TPK3-YFP-, and TPK5-YFP-expressing cells clearly originated from outside the plastid, likely from the tonoplast, whereas KEA3-YFP fluorescence was observed exclusively in leaf plastids. Scale bar = 50 μm.
KEA3 But Not TPK3 Is Preferentially Expressed in Photosynthetically Active Leaf Tissue
To gain insight into the tissue-specific gene expression of TPK3, we carried out a GUS-reporter assay using a previously described TPK3 promoter GUS line (pTPK3::GUS; Voelker et al., 2006) side by side with a newly designed pKEA3::GUS transgenic reporter line. Authenticity of the GUS stain was verified in wild-type plants (same developmental stage) as a negative control.
In agreement with previous work, GUS activity driven by the TPK3 promoter was found to be highest in seedling roots, cotyledons, and the stamina and pollen sacs of the flower tissue (Fig. 4). Although some signal was found in the youngest emerging true leaves, GUS stain was mostly absent from the adult leaf tissue and only found restricted to hydathodes (Fig. 4E). However, if the GUS reporter was expressed under control of the KEA3 promoter, the GUS staining was observed exclusively in aerial tissue, particularly near the vascular tissues of the leaves. The strongest signal was observed in seedling cotyledons and mature leaf tissue. In contrast to TPK3, no KEA3 expression was observed in flowers (Fig. 4G). The KEA3 expression pattern documented in this study confirms previous results (Han et al., 2015).
Figure 4.
TPK3 and KEA3 GUS reporter gene study in Arabidopsis tissues of different developmental stages. The following genes and stages were examined: pTPK3::GUS and pKEA3::GUS expression in 4-d-old seedlings (A), 11-d-old plants (B and C), rosettes of 19-d-old plants grown on one-half strength MS agar plates (D), mature leaves of 28-d-old plants (E), and flower tissue of 4.5-week-old plants grown in one-half strength MS hydroponics culture (F and G). Bars = 1 mm (A, B and G), 2 mm (C, E and F, left) and 5 mm (D and F, center and right).
In summary, our results indicate that KEA3 is preferentially expressed in photosynthetically active leaf tissue, whereas TPK3 shows highest expression in flowers and young emerging leaf tissue.
TPK3-Specific Immunoblots Confirm Protein Localization Outside Chloroplasts
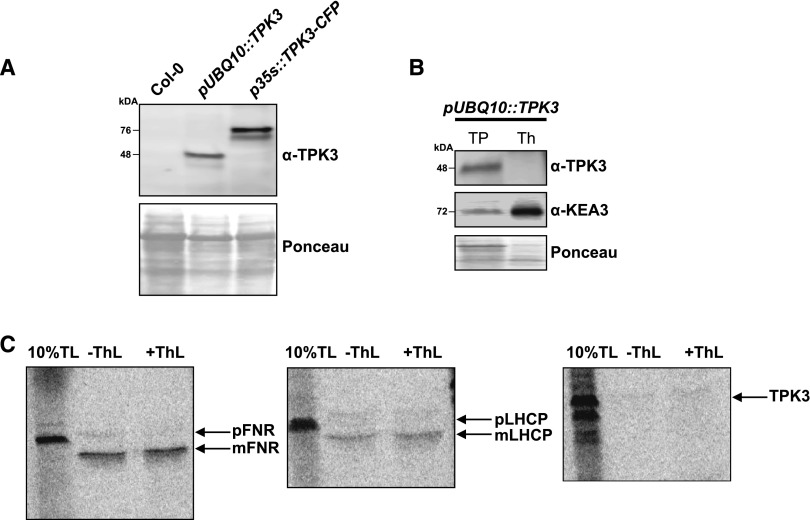
The presence of a fusion protein may affect in vivo protein targeting by masking the transit peptide. These effects can be hard to predict, especially for candidates that do not possess a classic plastid transit peptide, such as TPK3 (Armbruster et al., 2009). Thus, to verify our microscopy studies, we designed an Arabidopsis TPK3-specific antibody raised against an epitope located within the longest soluble loop in the center of the channel (Supplemental Fig. S3A). α-TPK3 was applied in immunoblots on total protein extracts from leaf tissue. The previously published α-KEA3 served as a thylakoid control (Armbruster et al., 2014). Whereas KEA3, the thylakoid K+/H+ exchanger, was detectable in wild-type total leaf protein extracts, we were unable to detect a TPK3-specific signal (Fig. 5A). In line with in silico gene expression data (Supplemental Fig. S3, B and C; Winter et al., 2007), or the AtGenExpress Visualization Tool, our pTPK3::GUS studies at various developmental stages revealed low TPK3 expression compared to that of KEA3 in mature leaves (Fig. 4). In fact, TPK3 was preferentially expressed in flowers. However, when we artificially triggered TPK3 transcription in leaf tissue through the use of tag-free pUBQ10::TPK3 (Supplemental Fig. S3D) and p35s::TPK3-CFP, TPK3 protein was detectable. A clear shift in protein size was seen between tag-free TPK3 (∼48 kD) and TPK3-CFP-tagged overexpression lines (∼76 kD), confirming α-TPK3 functionality and accuracy (Fig. 5A).
Figure 5.
Biochemical studies confirm the absence of TPK3 in chloroplast membranes. A, α-TPK3 (this study) did not detect a low endogenous TPK3 level in wild-type leaves but confirmed TPK3 overexpression in leaf tissue of stable pUBQ10::TPK3 and p35s::TPK3-CFP lines. As shown by Ponceau staining, equal total protein amounts from 0.5 mg fresh weight extracts were loaded. B, Whereas TPK3 was readily detected in the total leaf protein fraction (TP) from pUBQ10::TPK3 plants, only KEA3, not TPK3, was present in isolated thylakoids from the same line. C, In vitro import studies with 10% of the 35S-labled translation mix (TL) for FNRL1, LHCP, and TPK3 proteins confirmed no uptake into isolated chloroplasts or processing of TPK3 (prefixes p and m indicate the premature and mature protein, respectively). Half of the uptake mix was treated with thermolysin (ThL) to get rid of surface-adhered proteins.
In a second experiment, we isolated thylakoid membranes and total leaf proteins from the same tag-free pUBQ10::TPK3 overexpressor plants and detected TPK3 or KEA3 with their respective specific antibodies. As shown in Figure 5B, both TPK3 and KEA3 were detected in whole leaf protein extracts. However, the TPK3 signal was absent from isolated thylakoids. In contrast, KEA3 was found strongly enriched in thylakoid versus total protein fractions.
35S-labeled TPK3 Protein Is Not Imported into Isolated Intact Chloroplasts
Even though we could not detect TPK3 in isolated thylakoid membranes from pUBQ10::TPK3 overexpressor lines (Fig. 5B) we cannot entirely rule out that minute amounts of TPK3 may become inserted into the thylakoid membrane by an unknown dual-targeting pathway between vacuole and plastids. To follow up on this possibility, we performed studies in which 35S-labeled TPK3 protein was tested for import into isolated intact chloroplasts. As positive chloroplast controls, the stromal ferredoxin-NADP(H) oxidoreductase FNRL1 and the light-harvesting complex II subunit (LHCP) were also 35S-labeled. Following this procedure, all three proteins were separately incubated with intact chloroplasts at 25°C. For each protein, one set of chloroplasts was afterward treated with thermolysin to remove proteins that only adhered to the chloroplast outer surface. In the case of FNRL1 and LHCP, genuine protein uptake into chloroplasts, as well as protein processing, i.e. cleavage of the transit peptide yielding the truncated mature protein, were documented. However, TPK3 did not show binding to the chloroplast, protein import into plastids, or protein processing (Fig. 5C).
We conclude that TPK3 does not possess a classic N-terminal plastid transit peptide recognized by the translocon machinery at the outer and inner chloroplast envelope membranes. Therefore, a hypothetical vacuole-plastid dual targeting mechanism would have to bypass the main protein import pathway. Such a mechanism would be surprising since >70% of the thylakoid membrane consists of monogalactosyldiacylglycerol and digalactosyldiacylglycerol (Dörmann and Benning, 2002), which provide a strikingly different lipid composition compared to that of the tonoplast (Zhang et al., 2015).
In summary, the results we obtained employing three independent localization techniques do not support the notion that TPK3 is present in the chloroplast or the thylakoid membrane. In addition, TPK3 appears to be preferentially expressed in flowers and young emerging leaves but not in photosynthetically active mature leaf tissue, unlike the thylakoid-localized K+ carrier KEA3. Altogether, our studies substantiate the absence of TPK3 in all publicly available (to the best of our knowledge) chloroplast proteomic datasets (Sun et al., 2009; Tomizioli et al., 2014; Hooper et al., 2017).
TPK3 Loss or Gain of Function Does Not Affect Photosynthetic Performance
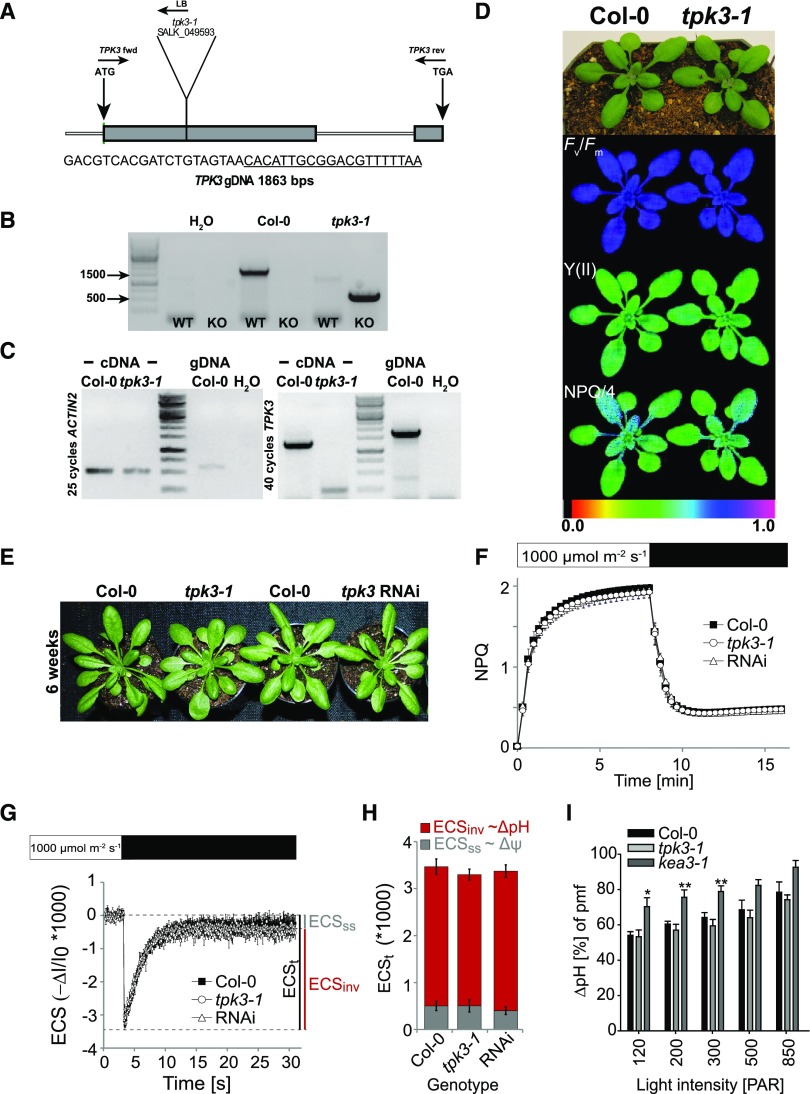
Since our experiments showed that TPK3 resides outside the chloroplast, we aimed to review its suggested function in regulating photosynthesis (Carraretto et al., 2013). Using The Arabidopsis Information Resource database, we found SALK_049593.1 to contain a T-DNA insertion in the first exon of the TPK3 locus (Fig. 6A). This specific TPK3 T-DNA insertion had been revealed only recently by reanalyzing all available T-DNA insertion lines using next generation sequencing (Jupe et al., 2019) and represents to our knowledge the only publicly available line with an insertion in the TPK3 coding region. Subsequently, we isolated homozygous tpk3-1 (SALK_049593) individuals by PCR and confirmed a T-DNA insertion in the first exon by sequencing (Fig. 6B). Full-length TPK3 transcripts were absent in tpk3-1 leaf tissue, as shown by RT-PCR, and 40 cycles were needed for detection in Col-0 (Fig. 6C), confirming low TPK3 expression in leaves (Fig. 4E). Under 16 h/8 h light/dark cycles at 150 µmol photons m−2 s−1, no visible abnormalities or changes in basic photosynthesis parameters between tpk3-1 and wild-type controls could be observed (Fig. 6D).
Figure 6.
tpk3-1 loss-of-function mutants and tpk3RNAi knock-down plants do not show a photosynthetic phenotype. A, Schematic representation of the T-DNA mutation in SALK_049593 inserted in the first exon of the TPK3 locus (the underlined sequence is the left border of the T-DNA). B, PCR on extracted genomic DNA showing homozygous TPK3 locus disruption in tpk3-1 individuals. C, Analysis of the TPK3 transcript level. Whereas ACTIN2 mRNA was detectable in both Col-0 and tpk3-1, TPK3 transcript was only found in the wild type after 40 PCR cycles. D, Truecolor and false color images of Col-0 and tpk3-1 of the photosynthetic parameters Fv/Fm, Y(II), and NPQ/4 grown in standard long-day conditions. E, No phenotypic alterations from Col-0 were found in tpk3RNAi and tpk3-1 loss-of-function lines when grown in the 12 h/12 h light/dark regime at 90 μmol photons m−2 s−1, as described in Carraretto et al. (2013). F to H, No differences between TPK3-deficient mutants and the wild type with respect to NPQ kinetics (F; means ± sd, n = 4), ECS relaxation after steady-state illumination (G), or pmf partitioning (H; means ± se, n ≥ 5). I, Increased pmf storage into ΔpH component was found upon light intensity ramping. Only kea3-1 plants had a significantly higher ΔpH component compared to that in Col-0 at 120, 200, and 300 μmol photons m−2 s−1 actinic light, as indicated with *P < 0.05 and **P < 0.01 using one-way ANOVA and the Holm-Sidak multiple comparisons test. The SE is based on n ≥ 8.
Next, Col-0, tpk3-1, and the previously published tpk3RNAi line no. 1 (Carraretto et al., 2013) were grown side by side in 12 h/12 h light/dark cycles at 90 µmol photons m−2 s−1, recapitulating the conditions previously reported (Carraretto et al., 2013). All genotypes grew without visible defects, i.e. no signs of anthocyanin accumulation were observed (Fig. 6E). Additionally, we were unable to reproduce the reported decreased NPQ induction at 1,000 µmol photons m−2 s−1 in the tpk3RNAi knock-down plants. NPQ induction in the tpk3-1 loss-of-function line also fully resembled wild-type NPQ kinetics (Fig. 6F). In Carraretto et al. (2013), the lower NPQ in the tpk3RNAi knock-down lines was correlated with a smaller pmf fraction stored as ΔpH. To probe pmf partitioning, we quantified the electrochromic shift (ECS) of the carotenoids, which provides a measure for the membrane potential (Δψ) across the thylakoid membrane (reviewed in Baker et al., 2007). ECS was monitored upon a dark transition in Col-0, tpk3RNAi, and tpk3-1 plants after reaching steady-state photosynthesis in the light (1,000 μmol photons m−2 s−1). In contrast to earlier reports, no changes in total pmf size or pmf partitioning from wild-type values were observed in tpk3RNAi and tpk3-1 plants (Fig. 6, G and H). Lastly, we tested how increasing actinic light intensities affect the distribution of ΔpH to total pmf in tpk3-1, and kea3-1 (Armbruster et al., 2014). Overall, with high light intensities, more pmf storage as ΔpH was observed (Fig. 6I). However, whereas loss of H+-dependent K+ exchange across the thylakoid membrane in kea3-1 resulted in a significantly increased pmf fraction stored as ΔpH at low to medium actinic light intensities, the ΔpH component in tpk3-1 remained at wild-type levels at any given light intensity (Fig. 6I).
Next, we probed the effects of TPK3 overexpression on photosynthesis. Initially, we transiently transformed N. benthamiana leaves followed by Chl a fluorescence analysis (Leonelli et al., 2016). Previously, transient expression of p35s::KEA3.3, encoding a KEA3 version lacking the regulatory C terminus, was shown to cause significant alterations in NPQ and PSII efficiency (ΦII), resembling the phenotypes recorded from respective stable p35s::KEA3 Arabidopsis overexpression lines (Armbruster et al., 2016). Whereas the effect of transient overexpression of p35s::KEA3.3-GFP reproduced lower NPQ and Fv/Fm values, leaf areas transformed with p35s::TPK3-CFP resembled those transformed with the p35s::TIC40-CFP negative control and untransformed leaf areas (Supplemental Fig. S4, A–D). A repetition of the experiment, with immunodetection for each construct, was performed in a second lab within our consortium and yielded the same results, i.e. no effect on NPQ by TPK3 overexpression (Supplemental Fig. S4, E and F). We validated this result by investigating two stable Arabidopsis overexpressor lines, pUBQ10::TPK3 and p35s::TPK3-CFP, grown under the conditions described by Carraretto et al. (2013). Again, no changes in NPQ or phenotypic alterations from that of Col-0 were detected (Supplemental Fig. S4, G and H).
Taken together, the results of our experiments show that neither gain nor loss of TPK3 function has an effect on plant health or affects photosynthesis under the tested conditions. These results are in line with a primary function for TPK3 in vacuoles of flower tissue (Fig. 4; Supplemental Fig. S3, B and C).
Recently, it was suggested that because of genetic redundancy potentially occurring among close TPK family members, posttranscriptionally silenced mutants, i.e. RNAi, represent a superior tool compared to T-DNA insertion lines (Szabò and Spetea, 2017). However, as in the case of TPK3, we also found its closest homologs, TPK2 and TPK5, to localize to the tonoplast (Fig. 3; Voelker et al., 2006) and thus, their primary function is most likely also unrelated to photosynthesis. Nevertheless, we wanted to exclude the possibility that the previously reported photosynthetic phenotype of the tpk3RNAi mutant was due to additional silencing of the close relatives TPK2 and TPK5. Thus, we isolated a tpk2tpk3tpk5 triple mutant. We also generated higher-order mutants with plastid members of the KEA family, i.e. thylakoid carrier KEA3 and the two envelope carriers KEA1 and KEA2, to test whether any unexpected functional relationship exists that may indicate a role for TPK3 in chloroplasts. However, when grown in 12 h/12 h light/dark cycles at 90 µmol photons m−2 s−1, none of the higher-order mutants with tpk3-1 showed any photosynthesis phenotypes different from that of their respective single or double mutants (Supplemental Fig. S5). Also, NPQ of tpk3-1 and tpk3-1kea3-1 resembled that of Col-0 and kea3-1, respectively, when analyzed under fluctuating irradiation (Supplemental Fig. S6, A–C). Lastly, we probed the leaf ionome of these mutants since the plastid ion transporter mutant kea1kea2 shows profound alterations (Höhner et al., 2016). However, besides the effects known for kea1kea2, no statistical significant alterations were observed for K, S, P, Ca, Mn, Fe, and Zn (Supplemental Fig. S6D). These results indicate that the effect of thylakoid K+ transport on the leaf elemental composition, such as by KEA3, is much more subtle compared to that of K+/H+ exchange across the envelope membrane.
Dynamic Environmental Photosynthetic Imaging Reveals Distinct Photosynthesis Impairments in Mutants Lacking Thylakoid Ion Transporters
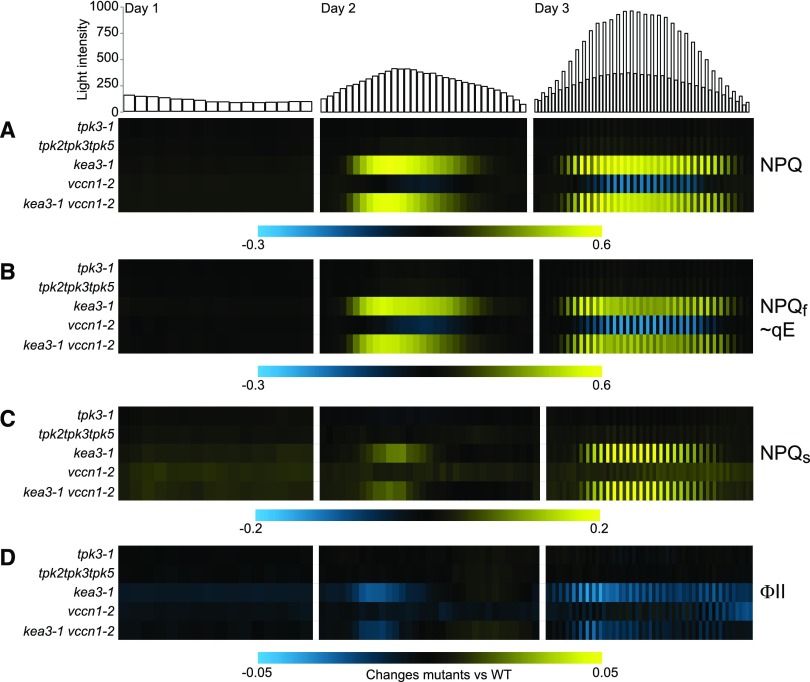
Previous work has shown that thylakoid ion transport is pivotal to adjust photosynthesis to rapid changes in light conditions. With the molecular nature of the thylakoid K+ channel still unknown, we sought to advance knowledge in the field by obtaining a comprehensive understanding of the roles of known thylakoid ion channel VCCN1 and K+ transporting H+ exchanger KEA3 (Armbruster et al., 2014; Herdean et al., 2016) in dynamic photosynthesis. For this purpose we systematically analyzed vccn1-2 and kea3-1 mutants and a newly isolated double mutant using Dynamic Environmental Photosynthetic Imaging (DEPI; Cruz et al., 2016). To test whether dynamic light-induced alterations also occur in tpk3-1 and tpk2tpk3tpk5 mutants, we added these mutants to the panel.
The DEPI run confirmed our previous observations in that tpk3-1 and tpk2tpk3tpk5 mutants behaved like Col-0, i.e. not significantly differently from the wild type, in all parameters and at all time points and light intensities tested (Fig. 7; Supplemental Dataset S1). These results provide additional support for a TPK3 (as well as TPK2 and TPK5) function outside of the thylakoid membrane and unrelated to photosynthesis regulation. Interestingly, the large-scale analysis of photosynthetic parameters by DEPI yielded new insights into the function of the thylakoid ion transport proteins KEA3 and VCCN1 and their interaction under dynamic light environments. Neither kea3-1 nor vccn1-2, nor their double mutant, differed from Col-0 with respect to their photosynthetic behavior when light intensity was supplied consistently at 150 µmol photons m−2 s−1 (day 1 of the DEPI set up). However, when exposed to dynamic light environments, such as a sinusoidal day (day 2) and a sinusoidal day plus superimposed high light phases (day 3), specific alterations in photosynthetic parameters became evident. The kea3-1 mutant displayed strongly increased NPQ nearly throughout days 2 and 3, except when light intensity was low at the beginning and end of the day (Fig. 7A; Supplemental Dataset S1). Most of the enhanced NPQ response in kea3-1 was due to increases in qE (Fig. 7B, NPQf), which is the fastest relaxing NPQ component and proportional to the proton concentration in the thylakoid lumen (Briantais et al., 1979). However, during high-light phases also NPQ components with slower relaxation kinetics than qE, i.e. NPQs, were strongly increased in kea3-1 compared to that in Col-0 (Fig. 7C). The composition of the individual NPQ components that form NPQs cannot be resolved in the DEPI set-up (Cruz et al., 2016). However, the pattern of the NPQs increase in kea3-1, with NPQs build-up only during the high-light phases but wild-type-like NPQs during the low-light phases, strongly argues against a marked contribution from the photoinhibitory NPQs component (qI). It takes qI much longer to relax than the 15 min that lie between the high- and low-light phases in the DEPI setup (Jahns and Holzwarth, 2012). Instead, the higher NPQs in kea3-1 may derive from increased zeaxanthin-dependent quenching component, a second NPQ component induced by low luminal pH (Nilkens et al., 2010). The elevated NPQ in kea3-1 during days 2 and 3 was accompanied by a decrease in PSII quantum efficiency, which was particularly pronounced during the peak light intensities of day 2 and day 3 (Fig. 7D; Supplemental Dataset S1).
Figure 7.
Unlike those in kea3 and vccn1 single and double mutants, photosynthetic parameters in tpk3-1 and higher-order tpk loss-of-function mutants remained at the wild-type level in DEPI runs on three representative days. NPQ (A), NPQf (B; NPQf ∼ qE), NPQs (C), and ΦII (D) are shown as changes from the wild type. No deviation in these parameters was detected in tpk3-1 or tpk2tpk3tpk5 triple-mutant plants. More information on the number of plants per genotype and statistical analyses can be found in Supplemental Dataset S1.
In line with earlier results, vccn1-2 mutants were less capable of inducing qE and thus NPQ in response to increased light intensities (Duan et al., 2016; Spetea et al., 2017). This behavior became apparent on day 3 (sinusoidal fluctuating light conditions) when vccn1-2 displayed significantly decreased NPQ and qE during the high light pulses around midday (Fig. 7A; Supplemental Dataset S1). Additionally, the vccn1-2 mutant showed consistently high NPQs at the end of fluctuating-light day 3 that was significantly increased compared to that in Col-0 (Fig. 7C; Supplemental Dataset S1). The build-up of NPQs throughout the day and its presence under both low- and high-light periods (in contrast to NPQs in kea3-1) suggests that the elevated NPQs in vccn1-2 does indeed derive from the photoinhibitory component qI.
To probe the interplay of the known thylakoid K+ and Cl− transport proteins KEA3 and VCCN1, we also generated a kea3-1vccn1-2 double mutant. The obtained results show that the lack of K+/H+ exchange via KEA3 dominates levels of qE and NPQ, i.e. qE and NPQ values recorded during day 2 and day 3 of kea3-1vccn1-2 were significantly different only from the vccn1-2 single mutant, not from kea3-1 (Fig. 7, A and B; Supplemental Dataset S1). Similarly, the PSII quantum yield (ΦII) of kea3-1vccn1-2 resembled that of the kea3-1 single mutant (Fig. 7D; Supplemental Dataset S1).
DISCUSSION
In this study, we obtained new localization data using fluorescent protein fusions which confirm the initial studies on TPK3 and its closest homologs showing their exclusive localization to the tonoplast (Voelker et al., 2006; Dunkel et al., 2008; Maîtrejean et al., 2011). Even though we tested the same (p35s; Carraretto et al., 2013) and an alternative promoter (pUBQ10; Grefen et al., 2010), we were unable to observe any construct-specific fluorescence signal from the chloroplast, where we could only detect Chl a fluorescence. Our findings spur the question why a recent study reported the chloroplast thylakoid membranes as the main subcellular TPK3 location (Carraretto et al., 2013). Although this remains only speculative, it is possible that transient expression in Arabidopsis, which is less efficient than that in N. benthamiana (Waadt et al., 2014; Rosas-Díaz et al., 2017), did not result in high numbers of transformed leaf cells. Based on this assumption and the very low signal-to-noise ratio seen in micrographs by Carraretto et al. (2013), it is plausible that Chl a fluorescence and light scatter were misinterpreted as p35s::TPK3-dsRED signal from thylakoids. The fact that no tonoplast TPK3-dsRED signal can be seen in their images supports the notion that the investigated cells did not express the fusion protein, i.e. they remained untransformed, as strong tonoplast TPK3 fluorescence protein signal has been observed in all our studies and those of others.
Additionally, our immunoblotting results clearly support the TPK3 protein localization outside the chloroplast. However, we will attempt to explain the observed discrepancies with other recent studies (Zanetti et al., 2010; Carraretto et al., 2013). Several reasons render it unlikely that the previously reported immunoglobulins/antisera were capable of precisely detecting the TPK3 channel in Arabidopsis. Whereas our TPK3 antibody was specifically designed to bind a TPK3 epitope, in previous studies antibodies were applied that were not directly or exclusively targeting TPK3 (αSynK, anti-K-PORE, and anti-TPK3/5; Supplemental Fig. S7, A and B); (1) αSynK is aimed against a 144 amino acid stretch of the K+ channel SynK (slr0498; Zanetti et al., 2010), in which the sequence similarity to TPK3 is particularly low (Supplemental Fig. S7A); (2) Anti-K-PORE is targeted against a K+ channel pore domain (pfam07885; Zanetti et al., 2010) that is much more specific to potassium channel (AKT2; AT4G22200; 9 of 10 amino acids match) and TPK1 (AT5G55630; 8 of 10 amino acids match) than to TPK3 (6 of 10 amino acids match; Supplemental Fig. S7B). The pfam07885 domain is shared by at least 15 members spread across the AKT and TPK family in Arabidopsis (Schwacke et al., 2003). It is therefore questionable whether the anti-K-PORE immunoglobulin would possess sufficient affinity to preferably bind and detect the TPK3 channel that is barely expressed in leaf tissue (Fig. 4, GUS data; Supplemental Fig. S3, B and C) rather than the more highly expressed AKT2 and TPK1 (Schmid et al., 2005), which also share higher sequence similarity with the epitope; and (3) The initially designated 3A8 monoclonal antibody (or anti-TPK3/5 antibody; Zanetti et al., 2010), later renamed monoclonal anti-TPK3 (Carraretto et al., 2013), was raised against a TPK5 antigen located in the short loop connecting transmembrane domains 1 and 2 (Supplemental Fig. S7B). In this stretch, TPK3 shares only 8 of 14 nonconsecutive amino acids. In conjunction with the results obtained using our own α-TPK3, we remain cautious regarding the feasibility of specifically detecting TPK3 in wild-type thylakoid fractions with any of the three different antibodies (αSynK, anti-K-PORE, or anti-TPK3/5) reported earlier.
In line with TPK3 localization in the vacuolar membrane, none of our chlorophyll fluorescence or pigment absorption-based analyses revealed any signs of compromised photosynthesis in the T-DNA insertion null mutant tpk3-1 isolated in this study. Moreover, further crossings with previously identified chloroplast ion transport mutants did not yield any phenotypic changes. Additionally, we did not find evidence for functional redundancy among TPK3 and its close relatives TPK2 and TPK5, which are also localized to the tonoplast. Even tpk2tpk3tpk5 triple mutants performed indistinguishably from wild-type control plants. Lastly, the original tpk3RNAi line also failed to reproduce earlier reported phenotypic abnormalities in our hands. In general, it is unclear how the tpk3RNAi construct employed by Carraretto et al. (2013) and Allorent et al. (2018) that was driven by the phloem-specific root locus C (rolC) promoter from Agrobacterium rhizogenes strain A4 (Molesini et al., 2009; Carraretto et al., 2013) should effectively silence TPK3 in photosynthetically active mesophyll cells. The employed root locus C promoter was shown previously to drive expression in plant phloem cells, but expression is absent in both epidermal and mesophyll cells (Yokoyama et al., 1994; Pandolfini et al., 2003). In line with RNAi’s proven function in tissue-specific posttranscriptional gene silencing (Byzova et al., 2004; Davuluri et al., 2005), silencing of TPK3 should thus be limited to phloem cells in tpk3RNAi lines.
Whereas we demonstrate that TPK3 does not have a function in photosynthesis, our long-term phenotyping results from dynamic light environments underline that two thylakoid ion-transport proteins, the K+/H+ antiporter KEA3 and the Cl− channel VCCN1, are necessary for optimal photosynthesis specifically under such conditions (Fig. 7). Loss of KEA3 leads to a strongly increased photoprotective response, which is accompanied by a decrease in the quantum efficiency of PSII. These results lead us to hypothesize that a thylakoid K+ channel, which counteracts KEA3-driven K+ import into the lumen, should similarly impact photoprotection and photosynthetic efficiency in a positive manner. The presence of K+ channel activity in the thylakoid membrane has long been reported (Hind et al., 1974; Tester and Blatt, 1989) and the importance of a K+ channel for photosynthesis has recently been corroborated by a computational model of transthylakoid ion flux and its impact on photosynthesis (Davis et al., 2017). Furthermore, the significant increase of qI in vccn1-2 as a consequence of dynamic light supports the idea that, upon increases in light intensity, Δψ dissipation by thylakoid ion channels (e.g. VCCN1) plays a crucial role in avoiding PSII photoinhibition (Davis et al., 2016; reviewed by Armbruster et al., 2017). Therefore, we hypothesize that an additional important contribution of the elusive K+ channel is to protect PSII from Δψ-induced damage in fluctuating light.
CONCLUSION
Based on our experimental results, we conclude the following: (1) TPK3 is predominantly expressed in Arabidopsis flowers but not in leaf tissue. The physiological relevance of TPK3 in the flower requires further investigation; (2) TPK3 resides in the tonoplast, and there is no indication for its localization in the thylakoid membrane. Using independent approaches (fluorescence microcopy, immunoblotting, and in vitro protein uptake studies), we found no evidence to support the localization of TPK3 in the thylakoid membrane; (3) In line with a predominant vacuolar function for TPK3 and its two closest homologs, TPK2 and TPK5, we did not observe any changes in photosynthesis in respective single or deduced higher-order tpk null mutants; and (4) Our data from mutants defective in thylakoid K+ transport (kea3-1) and Cl− channel activity (vccn1-2) support an important role of a thylakoid-localized K+ channel for photosynthesis in dynamic light environments.
Much interest has been raised in adjusting NPQ dynamics to increase photosynthetic efficiency in plants (Kromdijk et al., 2016). Ion transport mechanisms in the chloroplast, and especially in the thylakoid membrane, could have a significant contribution to this innovative improvement strategy (Armbruster et al., 2017; Davis et al., 2017). However, we remain skeptical that TPK3 represents a useful target in this respect, since we found the K+ channel to only localize outside chloroplasts. Even if minute TPK3 amounts do insert into the thylakoid membrane, which we cannot entirely exclude, they do not exert any detectable effect on photosynthesis, as neither TPK3 loss nor TPK3 overexpression impacted photosynthetic parameters in our study.
MATERIALS AND METHODS
Plant Growth
Wild-type Arabidopsis (Arabidopsis thaliana) accession Col-0 and mutant plants were either germinated on one-half strength Murashige and Skoog (MS) 1% (w/v) phytoagar plates, pH 5.8, for 5 d, transferred to soil for 5 d in 150 µmol photons m−2 s−1 illumination and then transferred to 90 µmol photons m−2 s−1 illumination, or grown from the beginning at this last light intensity in 12 h/12 h day/night cycle at temperatures of 23°C and 21°C (in light and dark, respectively). Rosettes of 4- to 6-week-old plants were used for all experiments if not stated differently.
Plants used for the GUS promoter assays and total reflection x-ray fluorescence spectroscopy (TXRF) measurements were grown on one-half strength MS medium 1% (w/v) phytoagar plates or in one-quarter strength MS hydroponics cultures, respectively. To prepare seedlings for hydroponics, seeds were germinated in bottomless PCR tubes filled with one-half strength MS medium 0.8% (w/v) phytoagar until the root tip had penetrated the agar and emerged at the bottom of the tube. One-week-old plants in PCR tubes were then transferred to one-quarter strength MS medium and grown in a 16 h/8 h day/night cycle at 150 µmol quanta m−2 s−1 illumination. Four-week-old rosettes were harvested for elemental analysis by TXRF. For the GUS promoter assay, plants were harvested at diverse developmental stages (Fig. 4).
Isolation of Single and Higher-Order Mutants
tpk3-1 mutant (SALK_049593) was isolated by PCR using primers listed in the Supplemental Table S1. After three rounds of backcrossing followed by selfing of a heterozygous progeny, a homozygous seed pool from a single tpk3-1 individual was harvested and progenies were used in all physiological studies. Whole leaf rosette mRNA was isolated and transcribed into complementary DNA (cDNA). The cDNA pool was used to verify lack of TPK3 mRNA in tpk3-1 mutant. The tpk3-1 mutant was then crossed into kea3-1, kea1-1kea2-1, and tpk2-1tpk5-1 to generate higher-order mutants. tpk2-1 (SAIL_336_G09) and tpk5-1 (SALK_123690) single mutants were ordered from the stock center and confirmed via PCR. For genotyping primers, please see the Supplemental Table S1.
Generation of Stably Transformed Arabidopsis Plants
TPK3 overexpressing lines were generated by introducing the TPK3 coding sequence into the pGPTVII.CFP.Kan (p35s::TPK3-CFP; Waadt et al., 2008) and pHygIIUT-MCS (pUBQ10::TPK3) vectors (Kunz et al., 2014b). Each construct was transformed into Col-0 plants by floral dip (Clough and Bent, 1998). Individual transgenic plants were selected based on their resistance to Kanamycin (pGPTVII.CFP.Kan) and Hygromycin (pHygIIUT). Presence of the inserted TPK3 sequence was confirmed by PCR and its expression by protein immunoblotting.
Similarly, pKEA3::GUS plants were generated by fusing the 5′ upstream region containing the KEA3 promoter (1.85 kb) to the β-glucuronidase reporter gene followed by the 3′ KEA3 genomic region (886 bp) and insertion into the pEarleyGate100 (pEG100) vector. The construct was introduced into Col-0 plants as mentioned above, and transgenic plants were selected based on their resistance to Basta and confirmed by PCR analysis.
Confocal Microscopy for Protein Localization Studies
Images were taken with a Leica SP8 Confocal Laser Scanning Microscope equipped with a supercontinuum laser. For colocalization experiments with YFP and chlorophyll the 514 nm line was used for excitation and emission was collected with hybrid detectors at 518–565 nm and 627–706 nm for YFP and chlorophyll autofluorescence, respectively. For colocalization analysis of YFP and mCherry, sequential frame scanning was conducted to prevent cross talk. YFP was excited at 514 nm and emitted light was collected at 518–565 nm. mCherry excitation occurred at 587 nm and emission at 594–671 nm was detected. In the case of colocalization analysis of CFP and YFP (Fig. 2), again sequential frame scanning was conducted. CFP was excited at 405 nm and emission signal was collected at 465–492 nm. Subsequently YFP was excited at 514 nm and light emission was collected at 518–565 nm.
GUS Staining
GUS staining using 5-bromo-4-chloro-3-indolyl β-D-glucuronide cyclohexylammonium salt (Chem Impex Inc.) was performed at several developmental stages. Plants were either grown on one-half strength MS plates or in hydroponics. For details on plant and tissue age, please see the Figure 4 legend. Plant tissue was stained in 100 mm phosphate buffer, pH 7.0, with Triton and 0.01% (w/v) 5-bromo-4-chloro-3-indolyl β-D-glucuronide cyclohexylammonium salt for 24 h at 37°C in the dark (Jefferson et al., 1987; Kosugi et al., 1990). To visualize the blue GUS staining, the leaf chlorophyll was extracted using a series of ethanol dilutions spanning 40% to 100% (v/v) ethanol.
Chlorophyll Quantification
Individual rosettes of wild-type or mutant plants were ground up in liquid nitrogen and 5–10 mg of frozen plant material was transferred into prechilled reaction tubes. After addition of 1 mL 80% (v/v) acetone, samples were incubated for at least 30 min on ice in the dark interrupted by occasional vortexing. Samples were centrifuged at 7,500 rpm for 5 min to pellet debris. Chlorophyll determination normalized to fresh weight per plant was performed on the clear supernatant (Porra et al., 1989).
Electrochromic Shift Measurements
To determine the partitioning of pmf, we took an approach using the ECS around 520 nm (reviewed in Baker et al., 2007; Bailleul et al., 2010). All measurements were performed on a custom-made spectrophotometer (Hall et al., 2013). Measuring wavelengths were supplied by LEDs with 5-nm bandpass filtering.
Wild-type and mutant plants were dark adapted for 30 min, then illuminated with 1,000 μmol photons m−2 s−1 actinic light for 10 min to reach steady state (Fig. 6G). For this data set, the dark interval relaxation kinetics (DIRK) for three wavelengths (505, 520, and 535 nm) were monitored and the ECS signal was deconvoluted as described in Cruz et al. (2001).
For the measurements at multiple actinic light intensities (Fig. 6I), leaves were preilluminated for 8 min at 100 μmol photons m−2 s−1 actinic light and 2 min before each DIRK measurement at the respective actinic light intensity of 120–850 μmol photons m−2 s−1. For these measurements, DIRK of the difference between 520 nm and 550 nm was monitored as described in Klughammer et al. (2013). Measurements were compared in the direction of increasing light intensity followed immediately by the direction of decreasing light intensity. No hysteresis effect between the two directions could be observed. Thus, leaves were constantly illuminated and measurements were performed with decreasing light intensities for all genotypes. Based on chlorophyll determinations (Supplemental Fig. S6B), which showed no differences among all tested genotypes, no correction for chlorophyll content was performed.
Chl a Fluorescence Measurements
Chl a fluorescence measurements were performed using a Walz Imaging PAM (Walz GmbH) on plants dark-adapted for 30 min. The actinic light of 600 and 185 µmol photons m−2 s−1 was switched on for 5 min after a 36-s dark period (Supplemental Figs. S4H and S5, respectively). Saturation pulses (6,000 μmol photons m−2 s−1) were applied every 20 s. NPQ induction curves under consistent light (Fig. 6F) were performed using a Walz Dual-PAM-100 system. Plants were illuminated with 1000 μmol photons m−2 s−1 red actinic light for 8 min followed by 8 min relaxation in the dark. NPQ induction curves in a fluctuating light regime were performed using the same saturation pulse settings but applying pulses every 30–40 s and using an actinic light regime from 90 to 900 µmol photons m−2 s−1 (Supplemental Fig. S6C). Calculation of photosynthetic parameters (Fv/Fm, ΦII, NPQ) was done according to Baker (2008). At the Center for Advanced Algal and Plant Phenotyping at Michigan State University, homozygous loss-of-function lines were monitored for a period of 5 consecutive days (Fig. 7) using dynamic light intensities as described earlier (Cruz et al., 2016). DEPI parameters were calculated as described in Cruz et al. (2016), in which qE is also referred to as NPQf and qI as NPQs, respectively.
Transient Gene Expression in Nicotiana benthamiana for Localization or Photosynthesis Studies
TPK3 cDNA-pHygIIUT-Venus, TPK3 cDNA-pKAN(35s)-CFP, TPK3 cDNA-FLAG-tag C-terminus-pEG100, TPK2 cDNA-YFP-pEG100, TPK5 cDNA-YFP-pEG100, Kea3.1 gDNA-pHygIIUT-Venus (Kunz et al., 2014a), KEA3.3cDNA-pB7FWG2 (Armbruster et al., 2016), TIC40 cDNA-pBARIIUT-mCherry-Cterm, CaS cDNA-pBARIIUT-mCherry-Cterm, SLAC1-pBarIIUT-Venus, and mVenus-2xFyve-pBar-35s-NosT constructs were transformed into Agrobacterium tumefaciens strain GV 3101. The resulting colonies were grown in liquid media along with the gene silencing suppressor strain p19. All cultures were resuspended in activation buffer (10 mm MES, pH 5.6, 10 mm MgCl2, and 150 µm Acetosyringone; Indofine) and OD600 values of 0.3 for silencing suppressor strain p19 (Waadt and Kudla, 2008). Cells were incubated for 2 h at room temperature before the N. benthamiana leaf infiltration. On day 3 after infiltration, leaves were either used for subcellular localization studies using a Leica SP8 (Leica) or for measuring photosynthesis parameters. The latter measurements were performed on a Walz Imaging PAM as described in Armbruster et al. (2016).
Western Blot Analyses
Total protein was extracted from liquid nitrogen frozen leaf tissue (20 mg) using a lysing Matrix D (MP Biomedicals) as described in Armbruster et al. (2016). Proteins were extracted by addition of 100 μL of protein extraction buffer (200 mm Tris, pH 6.8, 8% [w/v] SDS, 40% glycerol, and 200 mm dithiothreitol). Samples were heated at 65°C for 10 min. Thylakoid membranes were isolated as described in Armbruster et al. (2014). Then, 10 µL of total protein extract and thylakoids corresponding to 3 µg total chlorophyll were separated by SDS PAGE on a TGX Pre-Cast gel (Biorad), blotted onto a nitrocellulose membrane as in Schwarz et al. (2015), and detected after incubation with the specific TPK3 antibody (1:100 dilution) using a C-DiGit Blot Scanner (LI-COR Biosciences).
TXRF
Elemental analysis via TXRF was generally performed as described in our earlier study (Höhner et al., 2016), with the following adjustments. Whole rosettes from hydroponics cultured plants were harvested and completely dried in an oven at 70°C. Subsequently, the leaf rosettes were ground and powderized with a fresh pipette tip. Then, 2–3 mg (dry weight) of ground plant material was boiled in 750 µL HNO3 (analytical grade) until tissue digestion was complete. From this solution, 100 µL were added to 1,000 µL Gallium internal standard (c = 99.8 µL*L−1) to a final volume of 1,100 µL. Of this mixture, 10 µL was spotted on the quartz carrier, dried down, and subsequently measured in a Bruker Picofox S2 TXRF instrument (Bruker AXSnano). Based on the Ga concentration in the sample solution, each elemental concentration was calculated in mg*g−1 dry weight.
In Vitro Protein Import Studies on Isolated Intact Chloroplasts
The coding sequence of a 1000-bp TPK3 cDNA fragment was cloned into the vector pGEM-5Zf(+) (Promega) under the control of the T7 promoter. Radiolabeled expression was carried out using the TNT Coupled T7 Reticulocyte Lysate System (Promega) in the presence of 35S‐Met. The coding regions of FNRL1 (P10933; preprotein, 40 kD; mature, ∼35 kD) and the LHCII subunit LHCP (P07371; preprotein, 28 kD; mature, ∼24 kD) from Pisum sativum were cloned into the vector pSP65 (Promega) under the control of the SP6 promoter. Transcription of linearized plasmids was carried out in the presence of SP6 polymerase. Translation was carried out using the Flexi Rabbit Reticulocyte Lysate System (Promega). The import reaction contained isolated intact pea chloroplasts (Waegemann and Soll, 1991) equivalent to 20 μg chlorophyll in 200 μL import buffer (330 mm sorbitol, 50 mm HEPES/KOH, pH 7.6, 3 mm MgCl2, 10 mm Met, 10 mm Cys, 20 mm potassium gluconate, 10 mm NaHCO3, 0.2% [w/v] BSA), 3 mm ATP, and 5 µL 35S‐labeled translation products. Import reactions were initiated by the addition of translation product and carried out for 15 min at 25°C. Intact chloroplasts were reisolated through centrifugation through a 40% (v/v) Percoll cushion. After washing, half of the reaction was incubated with 1.5 µg thermolysin for 20 min on ice to digest nonimported preproteins. Import reactions were separated by SDS-PAGE and radioactive products were detected by phosphor plate imaging.
Accession Numbers
Throughout this study the following genetic loci were either cloned or otherwise used in the investigation: KEA3 (At4g04850), TPK2 (At5g46370), TPK3 (At4g18160), TPK5 (At4g01840), TIC40 (At5g16620), CaS (AT5G23060), SLAC1 (At1g12480), ACTIN2 (AT3G18780). Furthermore, the following publically available T-DNA insertion lines were employed: kea3-1 (Gabi_170G09; Armbruster et al., 2014), tpk3-1 (SALK_049593), tpk2-1 (SAIL_336_G09), tpk5-1 (SALK_123690), npq4-1 (Li et al., 2000), kea1-1kea2-1 (SAIL_586_D02, SALK_045324; Kunz et al., 2014a), vccn1-2 aka atbest1‐2 (GK_796C09; Duan et al., 2016; Herdean et al., 2016).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Fluorescence signal line plots and isolation of p35s::TPK3-CFP overexpressor lines.
Supplemental Figure S2. TPK2, TPK3, and TPK5 together form one phylogenetic subclade within the TPK family.
Supplemental Figure S3. TPK3 and KEA3 tissue-specific gene expression.
Supplemental Figure S4. Transient expression of TPK3 in N. benthamiana leaves and overexpression in Arabidopsis does not affect photosynthetic parameters.
Supplemental Figure S5. tpk3-1 single and tpk3-dependent higher-order mutants do not show photosynthetic alterations from control plants under constant light conditions.
Supplemental Figure S6. NPQ dynamics of tpk3-1, kea3-1, and double mutants under short-term fluctuating light conditions.
Supplemental Figure S7. SynK-TPK3 and TPK family member alignments with marked antigen sequence.
Supplemental Table S1. Primer purpose, names, and DNA sequences used in this study.
Supplemental Dataset S1. P-values for all genotypes grown in the DEPI chamber for all time points and light intensities tested.
Supplemental Movie S1. Z-series of TPK3-YFP fusion proteins (shown in yellow) transiently expressed in N. benthamiana leaves localize to the outside of the chloroplast. Shown in blue TIC40-CFP and in red Chl a fluorescence.
Acknowledgments
We thank Dr. I. Szabó (University of Padua) for providing tpk3RNAi lines (Carraretto et al., 2013). Dr. K. Czempinski (University of Potsdam) kindly provided the pTPK3::GUS reporter line, and Dr. R. Waadt (University of Heidelberg) provided Slac1-Stop pBarIIUT-Venus and mVenus-2xFyve + pBar-35s-NosT. We thank L. Savage and D. Hall (Michigan State University) for outstanding support with DEPI experiments and J. Rescher (Max Planck Institute Golm) for technical support. We are grateful to Dr. K. Niyogi (University of California Berkeley) for funding α-TPK3 and to Drs. H. Kirchhoff and M. Knoblauch (Washington State University) for help with flash spec and microscopy, respectively. A. Lefors (Washington State University) assisted in isolation of tpk3-1. H.-H.K. is grateful for initial funding to launch this project through National Science Foundation grant MCB-1616236 to J.I. Schroeder at the University of California San Diego.
Footnotes
This work was supported by ERA-CAPS funding from the National Science Foundation and Deutsche Forschungsgemeinschaft (DFG) to D.M.K, H.-H.K., and U.A, a National Science Foundation Career Award benefitting R.H., C.V., and H.-H.K. (IOS-1553506 to H.-H.K.), a DFG research grant (AR 808/5-1 to U.A.), and additional funding from the DFG (SFB-TR 175, project B06 to B.B.).
Articles can be viewed without a subscription.
References
- Allorent G, Byrdin M, Carraretto L, Morosinotto T, Szabo I, Finazzi G (2018) Global spectroscopic analysis to study the regulation of the photosynthetic proton motive force: A critical reappraisal. Biochim Biophys Acta 1859:676–683 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Hertle A, Makarenko E, Zühlke J, Pribil M, Dietzmann A, Schliebner I, Aseeva E, Fenino E, Scharfenberg M, et al. (2009) Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol Plant 2: 1325–1335 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Carrillo LR, Venema K, Pavlovic L, Schmidtmann E, Kornfeld A, Jahns P, Berry JA, Kramer DM, Jonikas MC (2014) Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nat Commun 5: 5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Leonelli L, Correa Galvis V, Strand D, Quinn EH, Jonikas MC, Niyogi KK (2016) Regulation and levels of the thylakoid K+/H+ antiporter KEA3 shape the dynamic response of photosynthesis in fluctuating light. Plant Cell Physiol 57: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Correa Galvis V, Kunz H-H, Strand DD (2017) The regulation of the chloroplast proton motive force plays a key role for photosynthesis in fluctuating light. Curr Opin Plant Biol 37: 56–62 [DOI] [PubMed] [Google Scholar]
- Bailleul B, Cardol P, Breyton C, Finazzi G (2010) Electrochromism: A useful probe to study algal photosynthesis. Photosynth Res 106: 179–189 [DOI] [PubMed] [Google Scholar]
- Baker NR. (2008) Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu Rev Plant Biol 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Baker NR, Harbinson J, Kramer DM (2007) Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant Cell Environ 30: 1107–1125 [DOI] [PubMed] [Google Scholar]
- Briantais JM, Vernotte C, Picaud M, Krause GH (1979) A quantitative study of the slow decline of chlorophyll a fluorescence in isolated chloroplasts. Biochim Biophys Acta 548: 128–138 [DOI] [PubMed] [Google Scholar]
- Byzova M, Verduyn C, De Brouwer D, De Block M (2004) Transforming petals into sepaloid organs in Arabidopsis and oilseed rape: Implementation of the hairpin RNA-mediated gene silencing technology in an organ-specific manner. Planta 218: 379–387 [DOI] [PubMed] [Google Scholar]
- Carraretto L, Formentin E, Teardo E, Checchetto V, Tomizioli M, Morosinotto T, Giacometti GM, Finazzi G, Szabó I (2013) A thylakoid-located two-pore K+ channel controls photosynthetic light utilization in plants. Science 342: 114–118 [DOI] [PubMed] [Google Scholar]
- Checchetto V, Segalla A, Allorent G, La Rocca N, Leanza L, Giacometti GM, Uozumi N, Finazzi G, Bergantino E, Szabò I (2012) Thylakoid potassium channel is required for efficient photosynthesis in cyanobacteria. Proc Natl Acad Sci USA 109: 11043–11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cruz JA, Sacksteder CA, Kanazawa A, Kramer DM (2001) Contribution of electric field (Δψ) to steady-state transthylakoid proton motive force (pmf) in vitro and in vivo. Control of pmf parsing into Δψ and ΔpH by ionic strength. Biochemistry 40: 1226–1237 [DOI] [PubMed] [Google Scholar]
- Cruz JA, Savage LJ, Zegarac R, Hall CC, Satoh-Cruz M, Davis GA, Kovac WK, Chen J, Kramer DM (2016) Dynamic environmental photosynthetic imaging reveals emergent phenotypes. Cell Syst 2: 365–377 [DOI] [PubMed] [Google Scholar]
- Davis GA, Kanazawa A, Schöttler MA, Kohzuma K, Froehlich JE, Rutherford AW, Satoh-Cruz M, Minhas D, Tietz S, Dhingra A, et al. (2016) Limitations to photosynthesis by proton motive force-induced photosystem II photodamage. eLife 5: e16921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GA, Rutherford AW, Kramer DM (2017) Hacking the thylakoid proton motive force for improved photosynthesis: Modulating ion flux rates that control proton motive force partitioning into Δψ and ΔpH. Philos Trans R Soc Lond B Biol Sci 372: 20160381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, et al. (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23: 890–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Benning C (2002) Galactolipids rule in seed plants. Trends Plant Sci 7: 112–118 [DOI] [PubMed] [Google Scholar]
- Duan Z, Kong F, Zhang L, Li W, Zhang J, Peng L (2016) A bestrophin-like protein modulates the proton motive force across the thylakoid membrane in Arabidopsis. J Integr Plant Biol 58: 848–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel M, Latz A, Schumacher K, Müller T, Becker D, Hedrich R (2008) Targeting of vacuolar membrane localized members of the TPK channel family. Mol Plant 1: 938–949 [DOI] [PubMed] [Google Scholar]
- Grefen C, Donald N, Hashimoto K, Kudla J, Schumacher K, Blatt MR (2010) A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J 64: 355–365 [DOI] [PubMed] [Google Scholar]
- Hall CC, Cruz J, Wood M, Zegarac R, DeMars D, Carpenter J, Kanazawa A, Kramer D (2013) Photosynthetic Measurements with the Idea Spec: An Integrated Diode Emitter Array Spectrophotometer/Fluorometer. Springer, Berlin, Heidelberg, pp 184–188 [Google Scholar]
- Han L, Li JL, Wang L, Shi WM, Su YH (2015) Identification and localized expression of putative K+/H+ antiporter genes in Arabidopsis. Acta Physiol Plant 37: 101 [Google Scholar]
- Hedrich R. (2012) Ion channels in plants. Physiol Rev 92: 1777–1811 [DOI] [PubMed] [Google Scholar]
- Herdean A, Teardo E, Nilsson AK, Pfeil BE, Johansson ON, Ünnep R, Nagy G, Zsiros O, Dana S, Solymosi K, et al. (2016) A voltage-dependent chloride channel fine-tunes photosynthesis in plants. Nat Commun 7: 11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind G, Nakatani HY, Izawa S (1974) Light-dependent redistribution of ions in suspensions of chloroplast thylakoid membranes. Proc Natl Acad Sci USA 71: 1484–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhner R, Tabatabaei S, Kunz H-H, Fittschen U (2016) A rapid total reflection X-ray fluorescence protocol for micro analyses of ion profiles in Arabidopsis thaliana. Spectrochim Acta B 125: 159–167 [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH (2017) SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res 45(D1): D1064–D1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahns P, Holzwarth AR (2012) The role of the xanthophyll cycle and of lutein in photoprotection of photosystem II. Biochim Biophys Acta 1817: 182–193 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupe F, Rivkin AC, Michael TP, Zander M, Motley ST, Sandoval JP, Slotkin RK, Chen H, Castanon R, Nery JR, et al. (2019) The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet 15: e1007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer C, Siebke K, Schreiber U (2013) Continuous ECS-indicated recording of the proton-motive charge flux in leaves. Photosynth Res 117: 471–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y, Nakajima K, Arai Y (1990) An improved assay for β-glucuronidase in transformed cells: Methanol almost completely suppresses a putative endogenous β-glucuronidase activityc. Plant Sci 70: 133–140 [Google Scholar]
- Kromdijk J, Głowacka K, Leonelli L, Gabilly ST, Iwai M, Niyogi KK, Long SP (2016) Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science 354: 857–861 [DOI] [PubMed] [Google Scholar]
- Kunz HH, Gierth M, Herdean A, Satoh-Cruz M, Kramer DM, Spetea C, Schroeder JI (2014a) Plastidial transporters KEA1, -2, and -3 are essential for chloroplast osmoregulation, integrity, and pH regulation in Arabidopsis. Proc Natl Acad Sci USA 111: 7480–7485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz HH, Zamani-Nour S, Häusler RE, Ludewig K, Schroeder JI, Malinova I, Fettke J, Flügge UI, Gierth M (2014b) Loss of cytosolic phosphoglucose isomerase affects carbohydrate metabolism in leaves and is essential for fertility of Arabidopsis. Plant Physiol 166: 753–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonelli L, Erickson E, Lyska D, Niyogi KK (2016) Transient expression in Nicotiana benthamiana for rapid functional analysis of genes involved in non-photochemical quenching and carotenoid biosynthesis. Plant J 88: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-P, Björkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK (2000) A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Maîtrejean M, Wudick MM, Voelker C, Prinsi B, Mueller-Roeber B, Czempinski K, Pedrazzini E, Vitale A (2011) Assembly and sorting of the tonoplast potassium channel AtTPK1 and its turnover by internalization into the vacuole. Plant Physiol 156: 1783–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molesini B, Pandolfini T, Rotino GL, Dani V, Spena A (2009) Aucsia gene silencing causes parthenocarpic fruit development in tomato. Plant Physiol 149: 534–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchie EH, Niyogi KK (2011) Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol 155: 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilkens M, Kress E, Lambrev P, Miloslavina Y, Müller M, Holzwarth AR, Jahns P (2010) Identification of a slowly inducible zeaxanthin-dependent component of non-photochemical quenching of chlorophyll fluorescence generated under steady-state conditions in Arabidopsis. Biochim Biophys Acta 1797: 466–475 [DOI] [PubMed] [Google Scholar]
- Nomura H, Komori T, Kobori M, Nakahira Y, Shiina T (2008) Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J 53: 988–998 [DOI] [PubMed] [Google Scholar]
- Pandolfini T, Molesini B, Avesani L, Spena A, Polverari A (2003) Expression of self-complementary hairpin RNA under the control of the rolC promoter confers systemic disease resistance to plum pox virus without preventing local infection. BMC Biotechnol 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975: 384–394 [Google Scholar]
- Rosas-Díaz T, Cana-Quijada P, Amorim-Silva V, Botella MA, Lozano-Durán R, Bejarano ER (2017) Arabidopsis NahG plants as a suitable and efficient system for transient expression using Agrobacterium tumefaciens. Mol Plant 10: 353–356 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flügge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz N, Armbruster U, Iven T, Brückle L, Melzer M, Feussner I, Jahns P (2015) Tissue-specific accumulation and regulation of zeaxanthin epoxidase in Arabidopsis reflect the multiple functions of the enzyme in plastids. Plant Cell Physiol 56: 346–357 [DOI] [PubMed] [Google Scholar]
- Singh MK, Krüger F, Beckmann H, Brumm S, Vermeer JEM, Munnik T, Mayer U, Stierhof YD, Grefen C, Schumacher K, et al. (2014) Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr Biol 24: 1383–1389 [DOI] [PubMed] [Google Scholar]
- Spetea C, Herdean A, Allorent G, Carraretto L, Finazzi G, Szabo I (2017) An update on the regulation of photosynthesis by thylakoid ion channels and transporters in Arabidopsis. Physiol Plant 161: 16–27 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PDB, van Wijk KJ (2009) PPDB, the plant proteomics database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabò I, Spetea C (2017) Impact of the ion transportome of chloroplasts on the optimization of photosynthesis. J Exp Bot 68: 3115–3128 [DOI] [PubMed] [Google Scholar]
- Tester M, Blatt MR (1989) Direct measurement of K+ channels in thylakoid membranes by incorporation of vesicles into planar lipid bilayers. Plant Physiol 91: 249–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizioli M, Lazar C, Brugière S, Burger T, Salvi D, Gatto L, Moyet L, Breckels LM, Hesse A-M, Lilley KS, et al. (2014) Deciphering thylakoid sub-compartments using a mass spectrometry-based approach. Mol Cell Proteomics 13: 2147–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang Y-F, Nishimura N, Chan W-Y, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, et al. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainonen JP, Sakuragi Y, Stael S, Tikkanen M, Allahverdiyeva Y, Paakkarinen V, Aro E, Suorsa M, Scheller HV, Vener AV, et al. (2008) Light regulation of CaS, a novel phosphoprotein in the thylakoid membrane of Arabidopsis thaliana. FEBS J 275: 1767–1777 [DOI] [PubMed] [Google Scholar]
- Voelker C, Schmidt D, Mueller-Roeber B, Czempinski K (2006) Members of the Arabidopsis AtTPK/KCO family form homomeric vacuolar channels in planta. Plant J 48: 296–306 [DOI] [PubMed] [Google Scholar]
- Waadt R, Kudla J (2008). In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc 2008: pdb.prot4995. [DOI] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Waadt R, Schlücking K, Schroeder JI, Kudla J (2014) Protein fragment bimolecular fluorescence complementation analyses for the in vivo study of protein-protein interactions and cellular protein complex localizations. In Sanchez-Serrano JJ and Salinas J, eds, Arabidopsis Protocols. Humana Press, Totowa, NJ, pp 629–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Soll J (1991) Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J 1: 149–158 [Google Scholar]
- Weinl S, Held K, Schlücking K, Steinhorst L, Kuhlgert S, Hippler M, Kudla J (2008) A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol 179: 675–686 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Hirose T, Fujii N, Aspuria ET, Kato A, Uchimiya H (1994) The rolC promoter of Agrobacterium rhizogenes Ri plasmid is activated by sucrose in transgenic tobacco plants. Mol Gen Genet 244: 15–22 [DOI] [PubMed] [Google Scholar]
- Zanetti M, Teardo E, La Rocca N, Zulkifli L, Checchetto V, Shijuku T, Sato Y, Giacometti GM, Uozumi N, Bergantino E, et al. (2010) A novel potassium channel in photosynthetic cyanobacteria. PLoS One 5: e10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hicks GR, Raikhel NV (2015) Molecular composition of plant vacuoles: Important but less understood regulations and roles of tonoplast lipids. Plants (Basel) 4: 320–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XG, Ort DR, Whitmarsh J, Long SP (2004) The slow reversibility of photosystem II thermal energy dissipation on transfer from high to low light may cause large losses in carbon gain by crop canopies: A theoretical analysis. J Exp Bot 55: 1167–1175 [DOI] [PubMed] [Google Scholar]