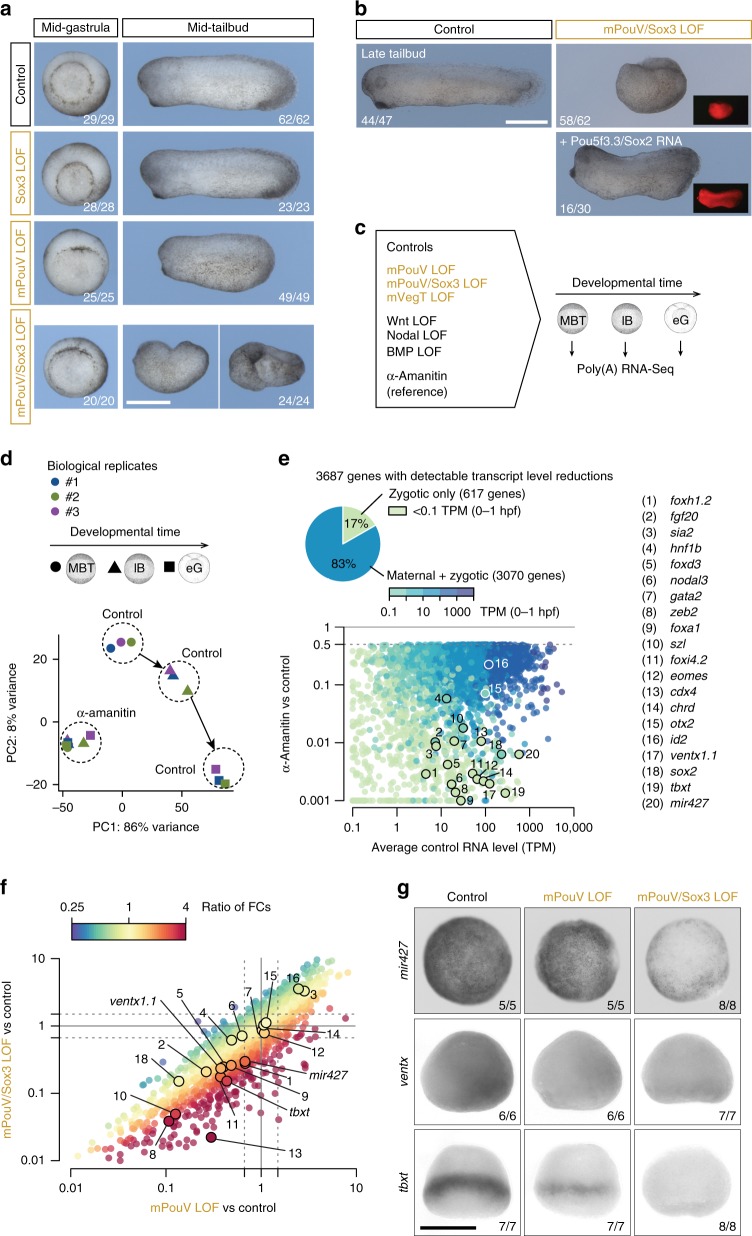
Fig. 6.
Synergistic relationship between maternal Pou5f3 (mPouV) and Sox3. a Morphological phenotypes caused by single and combined LOFs of Sox3 and mPouV when control embryos reached mid-gastrula and mid-tailbud stage. See also Supplementary Movie 1. b Phenotypical rescue of mPouV/Sox3 LOF embryos by the co-injection of both X. laevis Pou5f3.3 and Sox2 mRNA alongside mCherry mRNA as a tracer. c Experimental design: Profiling the poly(A) RNA transcriptome (n = 3 biologically independent samples) over three consecutive developmental stages under indicated conditions. Abbreviations used for the developmental timeline: MBT, midblastula transition; lB, late blastula; and eG, early gastrula. d Biplot of PC1 (accounting for 86% variance) and PC2 (8% variance) shows the relationship of developmental stage-specific poly(A) RNA transcriptomes of control and α-amanitin-injected embryos in biological triplicates (#1–3). e Detection of 3687 zygotic genes with reduced transcript levels (≥ 50%, FDR ≤10%) in α-amanitin-injected embryos. These genes were used as reference for all other LOFs. Dots in scatterplot are coloured according to the maternal contribution56 to the transcript level of each of these zygotic genes. f Scatterplot of transcript fold changes (FCs) caused by mPouV and mPouV/Sox3 LOFs with dots coloured according to the ratio of FCs. Numbered dots refer to some developmentally relevant genes listed in e. g Early gastrula-staged WMISH: mir427, animal view; ventx, lateral view, ventral side facing right; and tbxt, dorsal view. Numbers in the right bottom corner of each image refer to the count of embryos detected with the displayed WMISH staining among all embryos analysed per condition and in situ probe. Scale bars (a, b, g), 0.5 mm