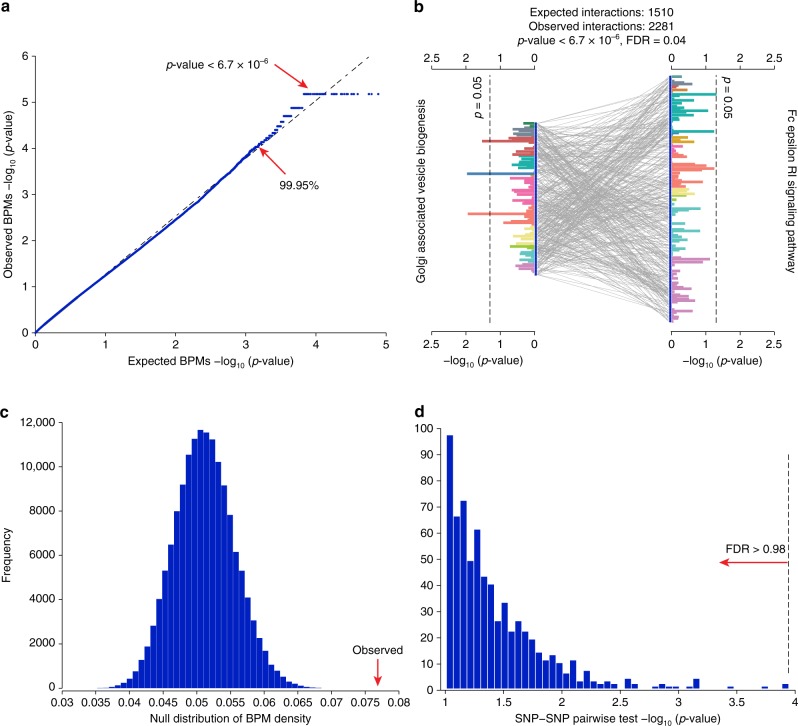
Fig. 2.
Significant pathway–pathway interactions discovered from the PD-NIA Parkinson’s disease cohort. a Quantile–quantile (QQ) plot comparing observed p-values (based on SNP-pathway membership permutations) for all possible pathway–pathway interactions between the 685 pathways to the expected, uniform distribution (log10 scale). The horizontal line at 6.7 × 10−6 reflects the maximum resolution supported by 150,000 permutations. b Interaction between Golgi-associated vesicle biogenesis pathway (Reactome) and FcεRI signaling pathway (KEGG). Two sets of SNPs mapped to genes in these pathways are connected by gray lines that reflect SNP–SNP interactions above a lenient top-5% percentile cutoff. The two groups of horizontal bars (grouped and colored by chromosome) show the −log10 p-values derived from a single-locus (univariate) test applied to each SNP individually (hypergeometric test), and the two dashed lines correspond to an uncorrected hypergeometric test p ≤ 0.05 cutoff, indicating that very few of the SNPs show marginal significant association before multiple hypothesis test correction. c Null distribution of the SNP–SNP interaction density between the Golgi-associated vesicle biogenesis pathway and FcεRI signaling pathway described in b based on 150,000 SNP permutations. The observed density for the Golgi-associated vesicle biogenesis and FcεRI signaling interaction is indicated by the red arrow and was not exceeded by any of the random instances (pperm < 6.7 × 10−6). d Distribution of p-values from individual tests for pairwise SNP–SNP interactions for SNP pairs supporting the pathway–pathway interaction, as measured by an additive disease model (−log10 p-value). None of the SNP pairs are significant after multiple hypothesis correction (dashed line at the most significant SNP–SNP pair corresponds to FDR = 0.98)