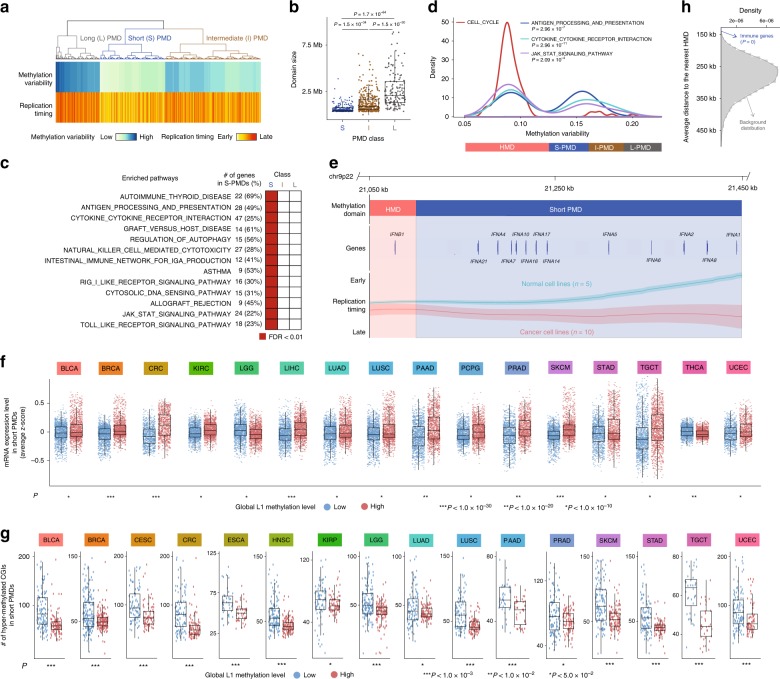
Fig. 4.
Characterization of genes in partially methylated domains. a Identification of PMD subclasses through hierarchical clustering on methylation variability and replication timing. b Comparison of PMD size between the identified PMD subclasses. c Enrichment of genes in immune-related pathways in the short PMDs. d Distribution of domain methylation variability for genes in cell cycle and immune pathways. The two-sample Kolmogorov-Smirnov test was used to assess deviation from the distribution of the cell cycle genes. e IFN-α genes in a short PMD with late replication timing. The mean and standard error of the weighted average signals of replication timing in normal cells and cancer cells are shown. f Comparison of the mRNA expression level of genes in the short PMDs between tumour samples with low and high global methylation. Tumour types for which the paired t-test P < 1.0 × 10−10 are shown. g Comparison of the number of hyper-methylated promoter CGIs in the short PMDs between tumour samples with low and high global methylation. Tumour types with P < 5.0 × 10−2 (two-sided Mann–Whitney U test) are shown. h Concentration of immune genes near PMD boundaries. The average distance of the immune-related genes (from b) to the nearest HMDs is marked by an arrow. The statistical significance of the observed average distance was assessed based on a null distribution generated by using random PMD genes