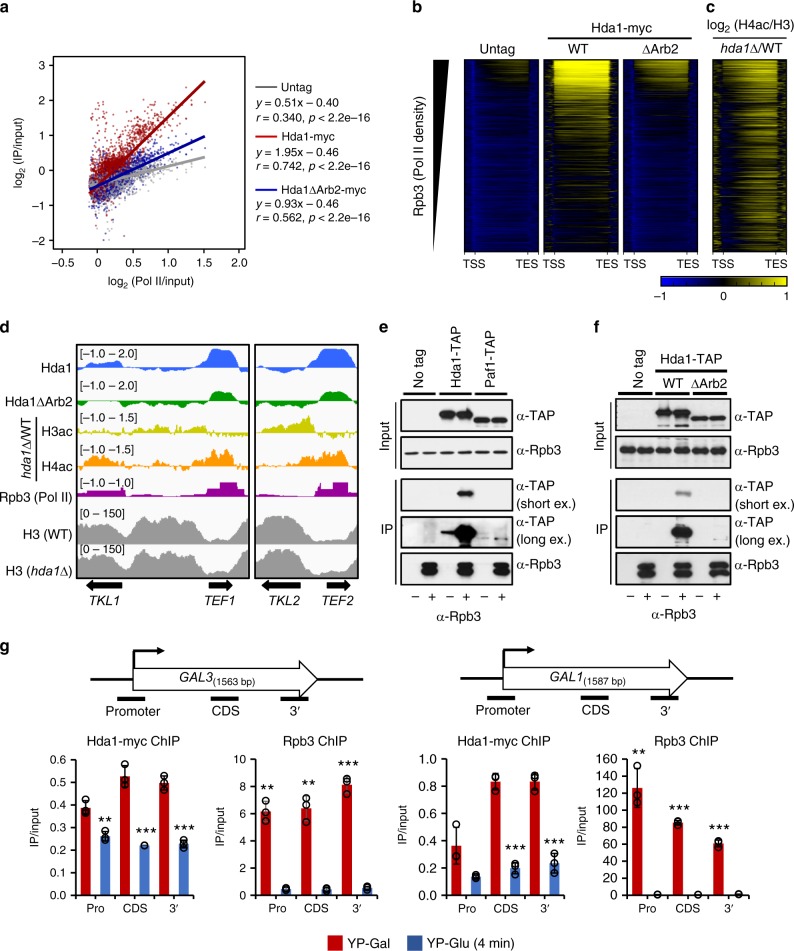
Fig. 4.
Transcription-dependent binding of Hda1C to active coding regions. a Hda1 binds to hyperactive genes. Scatterplot of Hda1 occupancy from two independent ChIP-seqs including S. pombe chromatin as spike-in controls in untagged (gray), Hda1-myc wild-type (red), and Hda1-myc (∆Arb2) (blue) cells, plotted against Rpb3 levels (relative Pol II). Pearson’s correlation coefficients are indicated. b Hda1 occupancies for the top 25% of highly transcribed genes (1705 genes) from two independent ChIP-seqs including S. pombe chromatin as spike-in control. The genes were sorted in descending order of Rpb3 occupancy in wild-type cells. The y axis indicates each gene and the x axis indicates relative position to the transcription start site (TSS) and transcription end site (TES). c Histone acetylation patterns of the genes shown in b. d ChIP-seq tracks at active genes, TKL1, TEF1, and TEF2, and an inactive gene, TKL2 showing the signals for Hda1 occupancy, H4 acetylation levels, H3 acetylation levels, Rpb3 occupancy, or histone H3 occupancy. e Hda1 interacts with Rpb3. Total extracts prepared from the indicated strains were incubated with an anti-Rpb3 antibody and protein G beads. The precipitates (IP) were analyzed by immunoblotting for TAP-tagged proteins (TAP) and Rpb3. Two independent experiments showed the same results. f The Arb2 domain is important for the interaction between Hda1 and Rpb3. Co-immunoprecipitation assay using the indicated strains was done as in e. Two independent experiments showed the same results. g Transcription-dependent Hda1C binding to GAL genes. The cells were grown in YP-Galactose (red) and then shifted to YP-Glucose for 4 min (blue). ChIP analyses of GAL3 and GAL1 were performed using anti-Rpb3 or anti-myc antibodies, as in Fig. 3a. Error bars show the standard deviation calculated from three biological replicates, each with three technical replicates. **p < 0.01 and ***p < 0.001 (two-tailed unpaired Student’s t tests). Source data are provided as a Source Data file