Figure 3. Sequential digestion of the 26S proteasome from Saccharomyces cerevisiae .
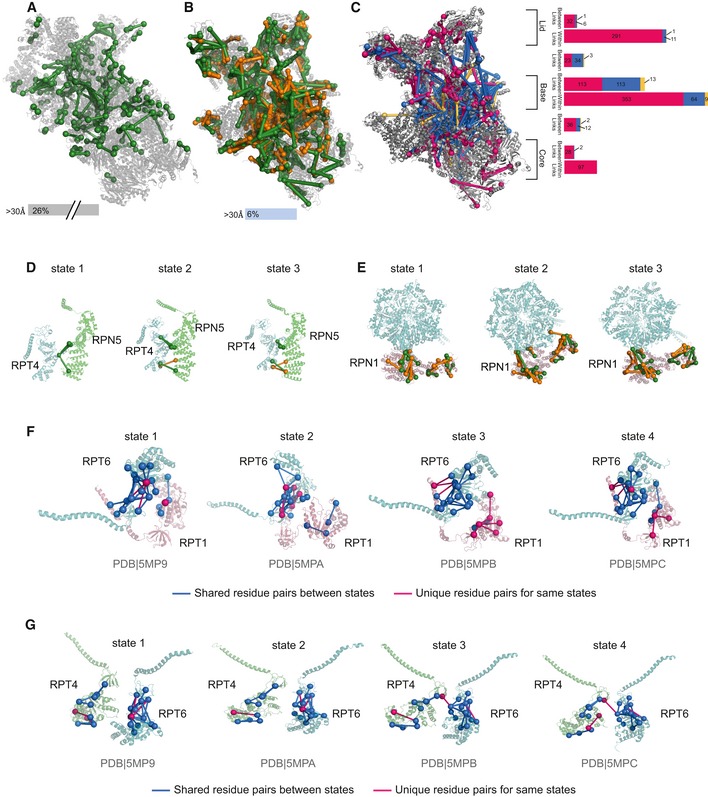
- Unique residue pairs obtained by Wang et al for the human 26S proteasome (PDB 5GJR).
- Unique residue pairs obtained by sequential digestion for the S. cerevisiae 26S proteasome (PDB 4CR2). Sequential digestion returned the highest number of residue pairs so far identified by CLMS for the 26S proteasome. Tryptic residue pairs are represented in green and non‐tryptic in orange.
- Long distance (blue) and within distance (pink) between residue pairs were mapped into one of the states of the proteasome (4CR2) showing the accumulation of those into the base of the complex. Residue pairs satisfying other states are represented in yellow. The bar plot shows the distribution of all residue pairs in the complex showing that long‐distance links locate mainly in the base.
- Unique residue pairs were mapped into the three states described by Unverdorben et al showing the rearrangement of Rpn5 relative to Rpt4.
- Our data support Rpn1 being translated and rotated to be positioned closer to the AAA‐ATPase.
- Structural rearrangements of the AAA‐ATPase‐dependent heterohexameric ring throughout four states for the RPT6 and RPT1 mapped to the four states described by Wehmer et al.
- Structural rearrangements of the AAA‐ATPase‐dependent heterohexameric ring throughout four states for the RPT4 and RPT6.
