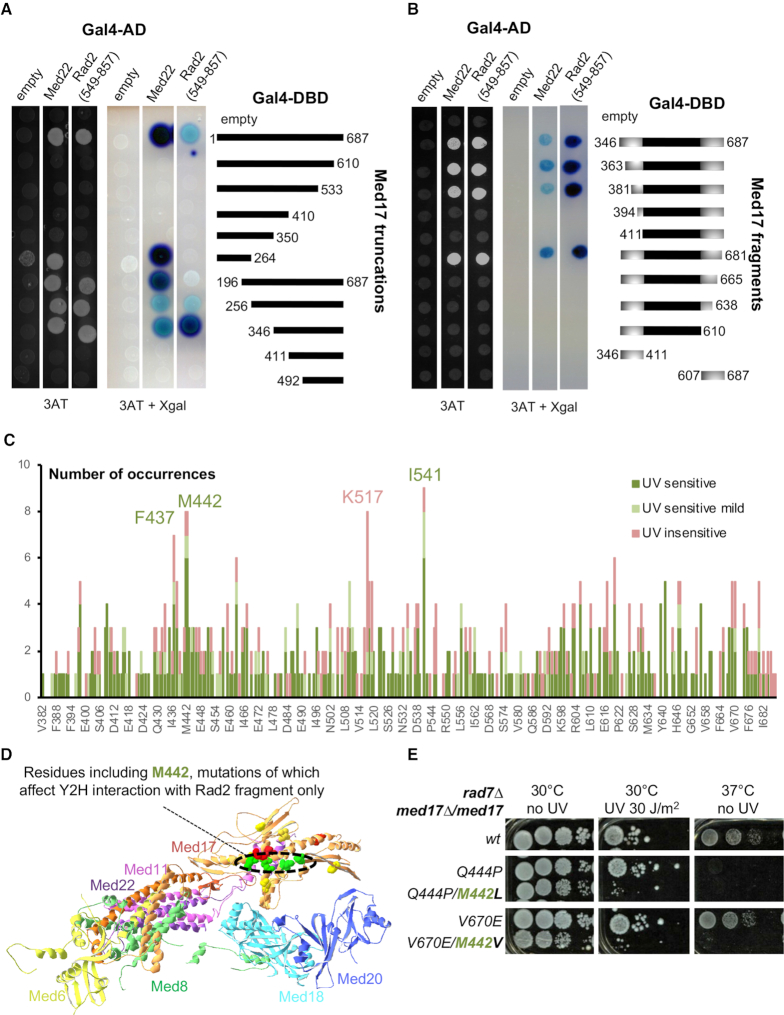
Figure 3.
Mutation analysis of Med17 region interacting with Rad2 fragment. (A andB) Serial truncations of Med17 were assayed for interaction with Med22 and a Rad2 fragment (549–857) identified in a yeast two-hybrid screening using Med17 as bait. (A) Gal4 DNA binding (Gal4-DBD) domain, alone (empty) or in fusion with full length Med17, 5 C-terminal truncations or 5 N-terminal truncations of Med17, was co-expressed with Gal4 Activation Domain (Gal4 AD), alone or in fusion with Med22 or residues 549 to 857 of Rad2. Yeast cells were spotted on SD+2A medium supplemented with 25 mM 3-AT, grown for 3 days (left panel) and then stained with X-Gal for 24h (middle panel). Residues 346–687 appear necessary and sufficient to have an interaction of Med17 with both Med22 and Rad2 fragment. (B) This minimal interaction domain was subjected to further N-terminal or C-terminal truncations, and N-terminal and C-terminal fragments of this domain were also tested for the interaction with Med22 and Rad2 fragment, using the same protocol as in panel A. (C) Pile histogram for occurrence of mutations affecting each residue in Med17 region interacting with Rad2, and the associated UV sensitivity phenotype. A graph for a complete Med17 protein is shown on Supplementary Figure S6D. The number of mutation occurrences is indicated for each amino acid position. Only mutants predicted to express full length Med17 were considered. The colours correspond to the associated phenotypes as follows: UV-sensitive in green, mild UV-sensitive in clear green and UV-insensitive in pink. More frequently mutated residues F437, M442, I541 were more frequently associated with UV sensitivity (in green), whereas K517 and nearby residues were more frequently found with temperature-sensitive only mutants (UV-insensitive in pink). (D) Med17 residues represented in Supplementary Figure S6E are highlighted in the structure of Mediator head module from Robinson and colleagues (PDB ID: 4GWP). Residues, mutations of which impaired or reduced Y2H Med17 interaction with Rad2 fragment specifically are in green (M442, K446, I541, N639). Residues, mutations of which impaired Y2H interaction of Med17 with Med22, Med11 and Rad2 fragment (K433, I445, I562) are in red. Residues, mutations of which impaired Y2H Med17 interaction with Med11 and Rad2 fragment, but not Med22, are in yellow (S409, V457, T509, I647) with the exception of M442. Other residues were not depicted. (E) Spotting assay to determine UV and temperature sensitivity of yeast strains expressing either WT or mutant versions of Med17 in rad7Δ context. Yeast cells were spotted on YPD agar plates, irradiated or not with 30 J/m2 UV-C (254 nm) and incubated at 30°C or 37°C for 3 days.