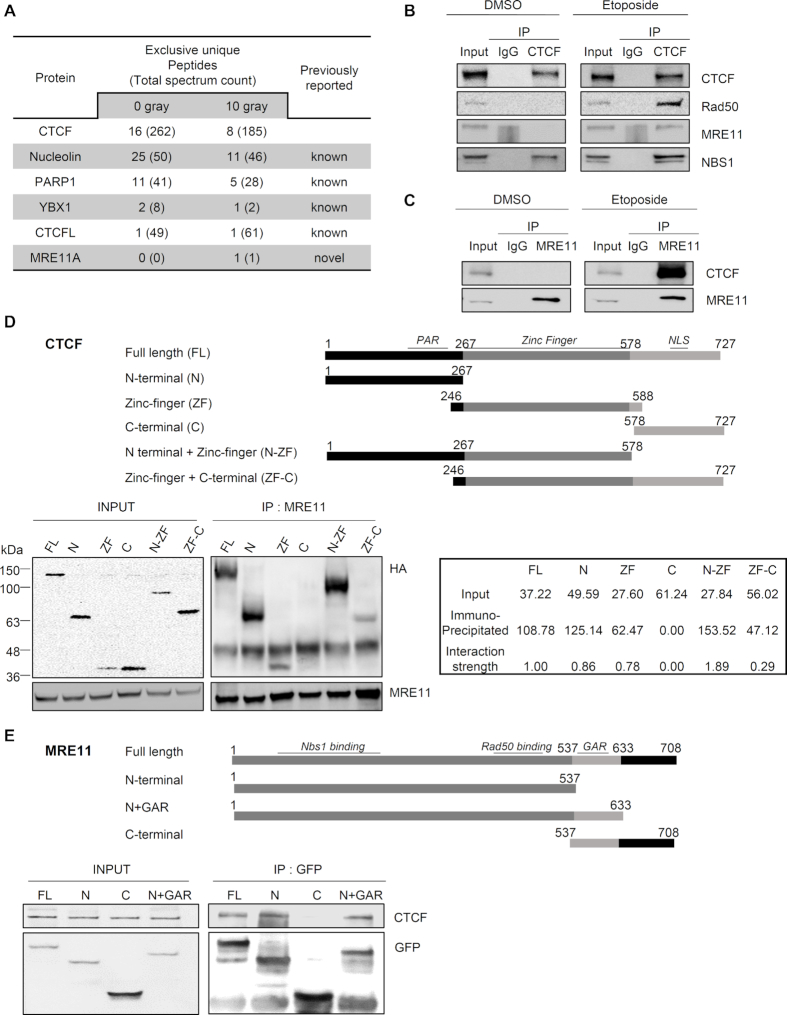
Figure 1.
CTCF interacts with MRE11 in response to DNA damage. (A) Selected protein list obtained from LC-MS/MS analysis after interactome tandem affinity purification of FLAG-SFB-tagged CTCF with or without γ-irradiation. The previously known and novel hits from MS results are shown. (B and C) Forward (B) and reciprocal (C) co-immunoprecipitation (co-IP) between endogenous CTCF and MRE11 in 293T cells, after the addition of DNase Benzonase, was performed with anti-CTCF (B) or anti-MRE11 (C) antibody, without (left, DMSO) and with (right, Etoposide) etoposide treatment. Immunoblot (IB) analysis was performed with the indicated antibodies. IgG immunoprecipitation (IP) was used as a negative control. See also Supplementary Figure S1. (D and E) Schematic representations of CTCF (D) and MRE11 (E) constructs used in this study (Top). The 293T cells were transfected with the indicated HA-tagged CTCF truncation constructs (D) or GFP-tagged MRE11 truncation constructs (E). Cell lysates were immunoprecipitated with anti-MRE11 (D) or anti-GFP (E) antibody, and immunoblot (IB) analyses were performed with the indicated antibodies. (D) Positions of molecular weight makers are denoted on the left of the INPUT SDS-PAGE gel. The interaction strengths between MRE11-HA and fusion CTCF proteins were normalized to the expression levels of HA fusion CTCF proteins, i.e. input. The interaction strength between MRE11-HA full-length CTCF was set as one, and the interaction strength of each HA fusion CTCF protein was calculated (bottom right panel); PAR, poly ADP-ribosylation; NLS, nuclear localization signal; N+GAR, N-terminus and glycine arginine rich motif.