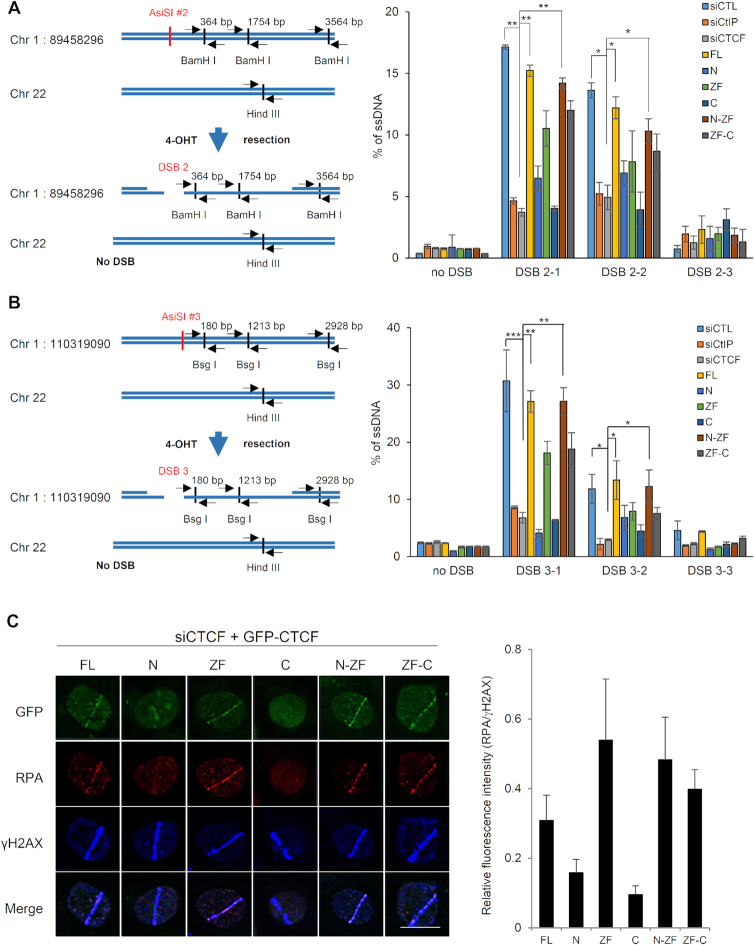
Figure 6.
CTCF depletion impairs DNA end resection. (A) Quantification of ssDNA generated by 5′-end resection at three sites (left, 2-1, 2-2, and 2–3 are 364, 1754 and 3564 bp from DSB, respectively and the paired primers across BamH 1 restriction sites are indicated as black arrow pairs) around the AsiSI-induced DSB (red) on chromosome 1:89,458,296 position in AsiSI–ER–U2OS cells transfected with the indicated shRNAs and HA-tagged CTCF constructs (right plot). The primers (black arrow pair) on chromosome 22 (no DSB) across a Hind III restriction site were used for negative control (left). Data are means ± SD of at least three independent experiments, and all qPCR reactions were performed in triplicate; *P≤ 0.05, **P≤ 0.01 (see Supplementary Figure S13A indicating levels of depleted CTCF or CtIP and complemented HA-tagged CTCF proteins.). (B) Design of qPCR primers and probes for measurement of resection at sites, 3-1 (180 bp), 3-2 (1213 bp) and 3-3 (2928 bp) near an AsiSI-induced DSB (red) on chromosome 1:110,319,090 position (left). Quantification of ssDNA generated from resection at three sites (left, 3-1, 3-2 and 3-3 located at Bsg I restriction sites) as in part (A) (right plot). Data represent means ± SEM of three independent experiments (see Supplementary Figure S13B indicating levels of silenced and complemented proteins.). (C) Accumulation of RPA onto the laser strips in CTCF-depleted U2OS cells complemented with the indicated GFP-tagged full-length or truncated CTCF proteins (left, see Figure 1A, above). Relative fluorescent intensities of RPA to those of γ-H2AX are presented in the right plot as the means ± SD of at least three independent experiments. More than 30 cells were counted in each experiment.