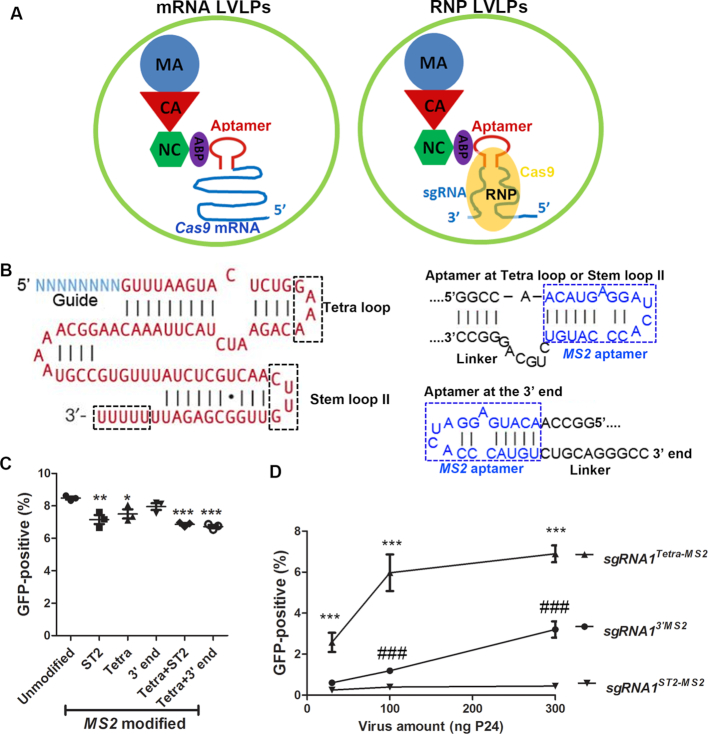
Figure 1.
Packaging sgRNA in LVLPs. (A) Diagram illustrating the difference between Cas9 mRNA LVLP (left) and Cas9/sgRNA RNP LVLP (right) as immature virion. Only one GAG precursor and one mRNA or RNP is shown for simplicity. ABP may bind to aptamer as dimers. Envelope proteins are not shown. (B) Locations for MS2 aptamer insertion in sgRNA scaffold. The original sgRNA is shown on the left. The ‘N’s indicate guide sequence. The three locations tested for MS2 aptamer insertion are indicated by the dashed black boxes. The inserted sequences are shown on the right. The blue letters in dashed blue boxes are the MS2 aptamers and the black letters are added linkers. Complementary ribonucleotides are indicated by vertical lines and atypical base-pairings are indicated by dots. (C) Effects of MS2 aptamer position on gene editing activity of the RNP with the modified sgRNA. Plasmid DNA co-expressing SaCas9 mRNA and various modified HBB sgRNA1 were transfected into the GFP-reporter cells and the GFP-positive percentage was determined by flow cytometry. Each data point indicates one independent experiment. *, ** and *** indicate P < 0.05, P < 0.01 and P < 0.001 (Tukey’s Multiple Comparison Test following ANOVA) when compared with unmodified HBB sgRNA1. (D) Effects of MS2 aptamer position on gene editing activity of modified sgRNA packaged in LVLPs. Indicated amounts of LVLPs containing MS2-modified HBB sgRNA1 and SaCas9 protein (or mRNA) were used to transduce 2.5 × 104 GFP reporter cells and the GFP-positive percentages were determined by flow cytometry. Each data point is the average of three replicates. ***, P < 0.001 when HBB sgRNA1TetraMS2 was compared with HBB sgRNA13′MS2; ###, P < 0.001 when HBB sgRNA13′MS2 was compared with HBB sgRNA1ST2MS2.