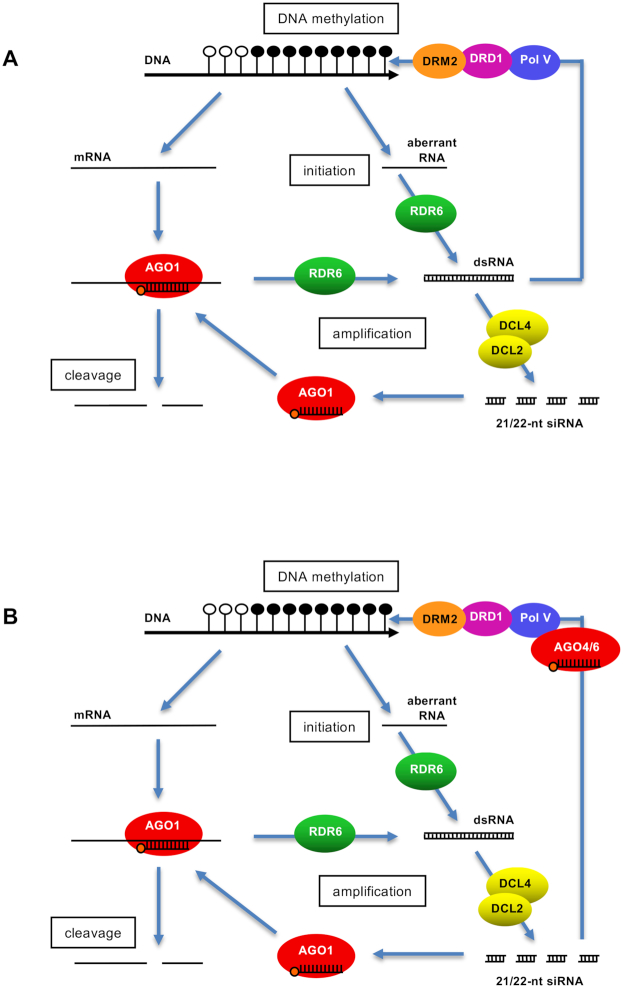
Figure 7.
Alternative models for PTGS-induced DNA methylation. S-PTGS is induced owing to the production of transgene aberrant RNAs that escape complete degradation by RQC pathways and are transformed into dsRNAs by RDR6, processed into primary 21–22-nt siRNAs by DCL2/DCL4, methylated by HEN1 and loaded onto AGO1 to guide the cleavage of regular mRNA. The binding of 22-nt siRNAs produced by DCL2 to mRNA may favor the recruitment of RDR6, leading to the production of secondary siRNAs and to amplifying the degradation process. To explain the pattern of DNA methylation in the transgene body, two hypotheses can be evoked: (A) RDR6-derived long dsRNAs directly trigger transgene DNA methylation through Pol V (NRPE1), DRD1 and DRM2, or (B) Part of the PTGS-derived siRNAs are loaded onto AGO4 and/or AGO6 and trigger transgene DNA methylation through Pol V (NRPE1), DRD1 and DRM2.