Abstract
Terminal deoxynucleotidyl transferase (TdT), which mediates template-independent polymerization with low specificity for nucleotides, has been used for nucleotide extension of DNA oligomers. One concern is that it is difficult to control the number of incorporated nucleotides, which is a limitation on the use of TdT for single-nucleotide incorporation of DNA oligomers. Herein, we uncovered an interesting inhibitory effect on TdT when ribonucleotide substrates (rNTPs) were employed in a borate buffer. On the basis of unique inhibitory effects of the ribonucleotide–borate complex, we developed a novel enzymatic method for single-nucleotide incorporation of a DNA oligomer with a modified rNTP by TdT. Single-nucleotide incorporation of a DNA oligomer with various modified rNTPs containing an oxanine, biotin, aminoallyl or N6-propargyl group was achieved with a high yield. The ‘TdT in rNTP-borate’ method is quite simple and efficient for preparing a single-nucleotide modified DNA oligomer, which is useful for the design of functional DNA-based systems.
INTRODUCTION
In materials science, the usefulness of oligodeoxynucleotides (ODN) has been proven in various applications requiring unique capacity for guided assembly, affinity, and catalysis (1,2). While researchers have endeavored to discover the value of DNA materials, the ODN modification technology has become their good companion. Nowadays, ODN modifications are regarded as an essential component of DNA-based sensors and nanoscale devices; these modifications provide functionality for detection, conjugation, and bioactivity of ODNs (3,4). The more DNA-based technologies progress, the more precise and desirable DNA modification tools (taking into account the flexibility of applications) are needed.
The enzymatic methods for ODN modification, such as those using a DNA polymerase, have been adopted in most relevant experiments because these methods provide a useful way of post-synthesis modification. Terminal deoxynucleotidyl Transferase (TdT) is one of the frequently used polymerases for modification of target DNAs. TdT possesses unique features that make it special among other polymerases. Because TdT has evolved as a specialized polymerase for nonhomologous end joining (NHEJ) (5–7), in which TdT participates in the DNA repair pathway by incorporating random nucleotides at the 3′-end of DNA fragments without a template strand. These two properties of TdT—the low substrate specificity for nucleotides and the template-independent polymerization—make the TdT-based method compatible with various kinds of modified nucleotides with convenient purification steps because there is no need to eliminate the template strand. Nonetheless, such template independence of TdT may be a double-edged sword for its applications in that its uncontrolled chain incorporation of nucleotides into an ODN happens due to the absence of a signal for termination. Until now, the use of TdT has been concentrated on the applications taking advantage of this uncontrolled incorporation of nucleotides, such as signal amplification (8,9) and polymerization of building blocks (10,11).
In this study, we would like to highlight a new use of TdT, that is, single-nucleotide incorporation by TdT; this approach has never been realized before. In the case of TdT, it is difficult to control the number of incorporated nucleotides, which is the limitation deterring the use of TdT for single-nucleotide modification of ODNs with a functional group. We started with the use of ribonucleotide triphosphates (rNTPs) as substrates instead of deoxyribonucleotide triphosphates (dNTPs; conventional substrates). There is evidence that TdT has a lower preference for rNTPs than dNTPs, and the use of rNTPs can be effective at reducing uncontrolled chain incorporation of nucleotides in a TdT reaction (12,13). Nevertheless, it still seems difficult to suppress this chain incorporation to achieve single-nucleotide modification of ODNs by TdT.
To develop a reliable method for single-nucleotide incorporation using TdT, which could be commonly applied to a wide range of functional nucleotides, we for the first time employed the rNTP–borate complex (Scheme 1A). The phenomenon where borate forms a reversible borate ester with cis-2′,3′-diol has been actively exploited in industrial chemistry (14). Only a few pioneering studies have pointed to the use in diagnostic and biosynthetic processes (15). In fact, the stability of the pentose–borate complex was studied, and it was reported that this complex has a positive effect on the stability of the RNA structure for life forms to develop at the primordial Earth (16–20), thus drawing scientific attention to the role of borate in a biological system. In the presence of the borate ion, ribonucleotide forms a ribonucleotide–borate complex, which is stable even at relatively low pH of 6.6 and high temperature (60°C) in saline (19,20). On the basis of the above information, we hypothesized that formation of the rNTP–borate complex would more effectively affect the chain nucleotide incorporation reaction of TdT. The process was simply implemented by carrying out a TdT reaction of rNTP in Tris-borate buffer (Scheme 1B). We found that the rNTP–borate complex can remarkably inhibit the chain elongation reaction of TdT. This result indicates that TdT could be used for single-nucleotide modification of ODNs with a modified nucleotide via the rNTP–borate complex.
Scheme 1.
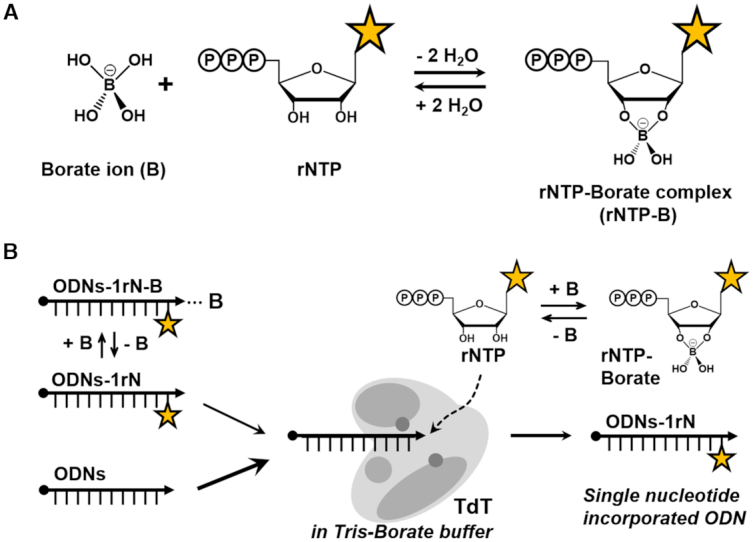
(A) The borate ion interacts with cis-diol (2′ and 3′ OH groups in ribose) of rNTP forming a rNTP–borate complex with a reversible borate ester. (B) A schematic diagram of the inhibitory pathway for rNTP incorporation in the TdT reaction influenced by Tris-borate buffer.
MATERIALS AND METHODS
Materials
Oligodeoxynucleotides (ODNs) were prepared by solid-phase chemical synthesis (Integrated DNA Technologies Co., Coralville, IA, USA). ODNs were used as follows: F-10dA (5′-/FAM/-AAAAAAAAAA) was employed for optimization of the TdT reaction, whereas F-20dN (5′-/FAM/-CTCAGGTCGACAGTCTGCGG, 20 nt) for ligation and conjugation testing. For the ligation assay, phosphorylated P19dN (5′-/Phos/-CCATTCCTGATTCTAAGTG, 19 nt) and T40dN (5′-CACTTAGAATCAGGAATGGCCCGCAGACTGTCGACCTGAG, 40 nt) were prepared as a downstream fragment and template oligomer, respectively. Three kinds of native nucleotide and four kinds of modified nucleotide were evaluated in this study. Each nucleotide substrate was as follows: GTP (cat. # G8877, Sigma-Aldrich, St. Louis, MO, USA), ATP (A1852, Sigma), UTP (U6625, Sigma), aminoallyl-UTP (AA-UTP) (cat. # R1091, Thermo Scientific™, Waltham, MA), N6-propargyl-ATP (N6P-ATP) (808490, Sigma), biotin-16-UTP (Bt-UTP) (11388908910, Roche, Basel, Switzerland). Oxanosine triphosphate (OTP) was manually synthesized from GTP. Terminal deoxynucleotidyl transferase (TdT) was purchased from New England Biolabs (M0315, NEB, Ipswich, MA). The cloned TdT gene was originated from calf thymus and the recombinant TdT was produced in Escherichia coli host system that carries the TdT expression vector. The purified TdT was supplied in 50 mM KPO4 (pH 7.3, 25°C), 100 mM NaCl, 1.43 mM 2-mercaptoethanol, 0.1% Triton X-100 and 50% glycerol. For the ligation assay, T4 DNA ligase (Promega, Madison, WI) was used. For conjugation testing, poly-l-lysine (Peptron Co., Daejeon, Korea) was purchased and applied as a reaction partner of OTP. Streptavidin (Thermo Scientific™, Waltham, MA, USA) was reacted with Bt-UTP. Sulfo-Cy5-NHS and sulfo-Cy5-azide dyes (Lumiprobe Co., Hunt Valley, MD, USA) were reacted with AA-UTP and N6P-ATP. All buffers used in this study were prepared with 2x reaction buffer which contained various concentration of borate; final 20 or 100 mM of Tris (T1503, Sigma-Aldrich) in combination with 0, 50, 100, 150 or 200 mM Borate (69025-0301, Junsei, Tokyo, Japan), pH 8.0. The solution of MgCl2 (100 mM, M4880, Sigma-Aldrich) and CoCl2 (2.5 mM, provided by NEB) was prepared to supply metal cofactors into the TdT reactions.
OTP preparation
The method of OTP preparation was taken from previous reports (21,22), which describes the synthesis of oxanine (Oxa) derivatives through deamination of guanine. Briefly, 10 mg of GTP was dissolved in 3.6 ml of 1 M acetate buffer (pH 3.7), then preheated for 5 min at 37°C. After the addition of 0.4 ml of 1 M NaNO2 (final concentration 100 mM) to the mixture, it was incubated for 4 h at 37°C. The mixture was neutralized by adding NaOH to stop the reaction. OTP was purified by HPLC (Young Lin Co., Korea, YL9100 system, Column: ULTRON VX-ODS 150 × 6.0 mm, 5 μm; gradient buffer system: 0% acetonitrile in 100 mM triethylammonium acetate (TEAA) at 0 min to 20% acetonitrile in 100 mM TEAA at 20 min, 1 ml/min flow rate) and then lyophilized.
TdT reaction with rNTP or modified rNTP for single-nucleotide incorporation
To obtain a high yield of a single-nucleotide-incorporated product, an optimized reaction of TdT in Tris-borate (TB) buffer was applied to each of the three normal nucleotides and four modified nucleotides. All components including the initiator (F-20dN, final concentration 3 μM), MgCl2 (final 10 mM), CoCl2 (final 0.25 mM), TdT (final 0.8 U/μl), and 2× TB buffer [final 100 mM Tris and 100 (or 150) mM borate, pH 8.0] were mixed at each optimal concentration of a modified nucleotide in distilled water (Supplementary Table S4). The optimal concentrations of normal or modified nucleotides are as follow: GTP (final 10 μM), ATP (final 30 μM) and UTP (final 100 μM); OTP (final 150 μM), N6P-ATP (final 200 μM), AA-UTP (final 50 μM) and Bt-UTP (final 200 μM), respectively. The mixture was incubated at 37°C for 60 min. To stop the reaction, 4 μl of 0.3 M EDTA was added per 25 μL of a reaction mixture, then heated at 90°C for 10 min. The reaction product was concentrated and rid of residual substrates by means of a 3 kDa MWCO centrifugal filter (Amicon™, Merck KGaA, Darmstadt, Germany).
Preparation of internally modified ODNs via ligation
Single-nucleotide incorporated ODN products (21 nt) that derived from a 20 nt initiator via a reaction of TdT in TB buffer were obtained as upstream fragments. The upstream fragment (final concentration 1 μM), a 5′ phosphorylated downstream fragment (P19dN, final 1.2 μM), and template (T40dN, final 1.2 μM) were incubated with T4 DNA ligase (3 U) in 100 μl of 1× a manufacturer-provided reaction buffer at 37°C for 3 h. The ligation product was purified on a desalting column, Centri-Sep™ (Thermo Fisher Scientific Inc., Waltham, MA, USA).
Conjugation
Each TdT product (21 nt) and ligation product (40 nt) was employed for conjugation with their binding or conjugation partner compound. The purified Oxa-modified ODNs (final 0.8 μM) and poly-l-lysine (final 300 μM) dissolved in water were mixed in 50 mM phosphate buffer pH 7.4 containing 150 mM NaCl (PBS), then it was incubated at 25°C for 48 h. The AA-modified ODNs (final concentration 1 μM) and Cy5-NHS (final 200 μM) dissolved in DMSO were added to 50 mM sodium carbonate buffer pH 9.0, then incubated at 25°C for 6 h. To carry out the conjugation between the N6P-modified ODNs and the Cy5-azide, we followed the copper-catalyzed alkyne-azide cycloaddition method with some modifications (31). The N6P-modified ODNs (final 1 μM) and Cy5-azide (final 10 μM) were added to a reaction buffer that consisted of 200 mM TEAA buffer pH 7.0, 1 mM ascorbic acid, and 30% DMSO. After addition of a 10 mM Cu-Tris(benzyltriazolylmethyl)amine (TBTA) solution (final 0.7 mM), the samples were incubated at 37°C for 1 day. These reaction products were separated by denaturing PAGE in a 12% gel (with 7 M urea). In case of biotin-modified ODNs, streptavidin was employed for affinity-based conjugation. The biotin-modified ODNs (final 0.8 μM) and streptavidin (final 10 μM) were mixed in PBS, and then incubated at 25°C for 30 min. The conjugate was separated by nondenaturing PAGE in an 8% gel at 4°C.
PAGE analysis and quantification
Reactants and reaction products were separated by denaturing PAGE in 7 M urea (16% gel) and quantified on a fluorescence image analyzer (LAS 4000, General Electric Co.) in the detection mode for the FAM dye (495 nm/520 nm). The concentration of incorporated dNTPs and rNTPs was calculated from the band intensity by regarding molar ratio of incorporated dNTPs and rNTPs.
RESULTS
Inhibitory effect of borate on incessant chain incorporation of ribonucleotides in TdT reaction
We prepared a set of Tris-borate (TB) buffers that contained different concentrations of borate. Although a 20 mM Tris-based buffer has been used generally for a normal TdT reaction, we adopted up to 100 mM Tris as the main buffer to find out the required concentration of borate so that this buffer serves as an effective chain incorporation regulator. The buffering range in each buffer was evaluated by titration (Supplementary Figure S1), showing that a high concentration of borate increased buffering capacity.
To determine whether the inhibitory effect of borate happened only to a ribonucleotide-type substrate, we compared the incorporation efficiency of substrates GTP (ribonucleotide substrate) and dGTP (deoxyribonucleotide substrate) in a TdT reaction in TB buffer (Figure 1A). The PAGE analysis indicated that incorporation efficiency of GTP into the 3′-end of ODNs was inversely proportional to the concentration of borate in TB buffer (lanes 2–5 in Figure 1A). In contrast, an influence of borate on the normal substrate, dGTP, was not observed (lanes 7–10 in Figure 1A). While deoxyribonucleotide substrate (dGTP) cannot interact with borate ion, ribonucleotide substrate (GTP) can make a cis-2′,3′-diol formation with borate, which might be critical for the inhibition on TdT elongation reactions. In a quantitative analysis performed on a set of TB buffers for various periods (Figure 1B), the concentration of incorporated GTP in 100:100 TB buffer (composed of 100 mM Tris and 100 mM borate) was 60% lower than that in 20 mM Tris buffer. Of note, slight inhibition was observed in the TdT reaction conducted in 100 mM Tris buffer. This phenomenon has been described in other reports: inhibition by a Tris buffer can be seen in a number of enzymatic reactions (lanes 5 and 10 in Figure 1A and B) (23,24). We confirmed that the inhibitory effect was not induced by a pH change, which may happen after the buffer dilution, but rather by the change in borate concentration (Supplementary Figure S2).
Figure 1.
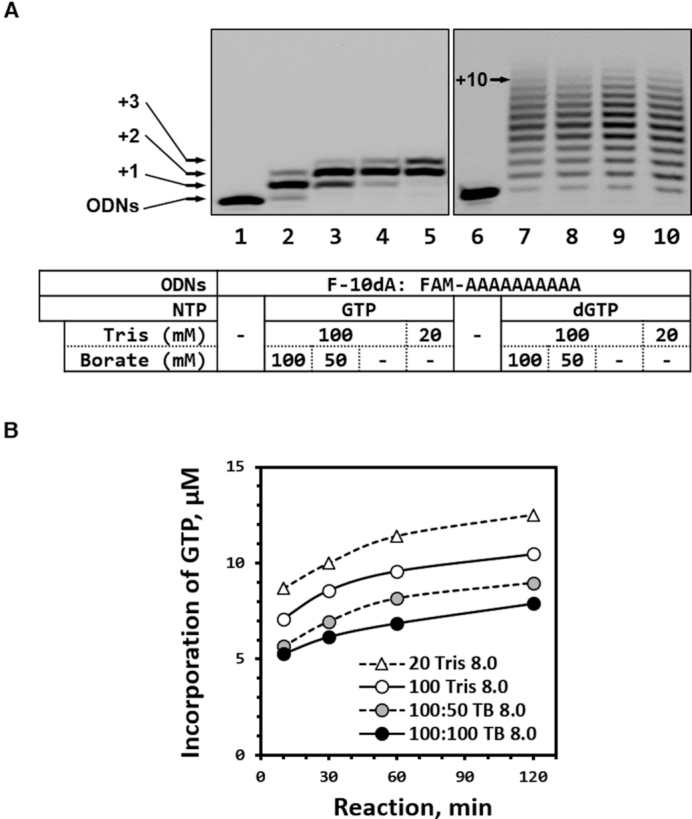
(A) Incorporation of GTP and dGTP into the initiator (F-10dA) depending on Tris and borate concentrations. (B) Time-course analysis of GTP incorporation at various borate concentrations in the Tris-based buffer, where 100:50 TB means that Tris-borate buffer is composed of 100 mM Tris and 50 mM borate in the working solution (1×). In brief, 30 μM GTP or 50 μM dGTP was applied to 3 μM initiator, which was labeled at the 5′-end with the FAM dye, with measurement of the incorporation efficiency by denaturing PAGE in a 16% gel (with 7 M urea).
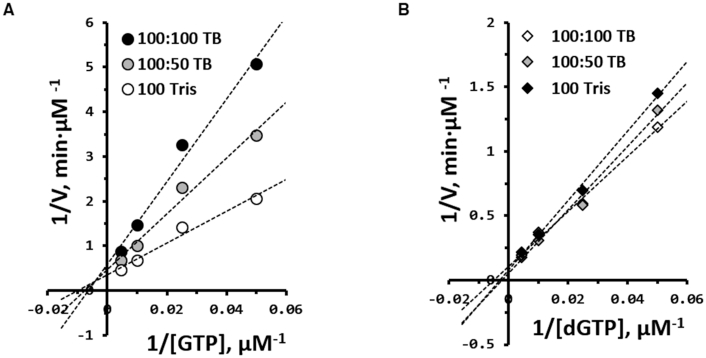
Enzyme kinetics evaluated with GTP in the set of TB buffers revealed the phenomenon of increased Km and decreased Kcat in accordance with the increased borate concentration; this phenomenon closely resembles a mixed inhibition pattern in a double reciprocal plot (Figure 2 and Supplementary Table S1). In contrast, no correlation between borate concentration and enzymatic reaction was detected when dGTP was used (Figure 2B). These results revealed that formation of the rNTP–borate complex (via borate esterification with cis-2′,3′-diol of rNTP) is the main factor causing the observed inhibition of chain incorporation by TdT (Scheme 1), and such inhibition is not caused by a direct interaction between the enzyme and borate. Compared with other competitive and noncompetitive inhibition mechanisms, the inhibitory mechanism of borate may be basically different, but its pseudo-mixed inhibition pattern can be explained as follows: Borate in solution binds to the 2′ and 3′ OH groups of rNTP to generate the rNTP–borate complex. The fact that increased borate concentration reduces the turnover rate of TdT probably means that this complexation indirectly interferes with the formation of a tertiary complex of the substrate with the enzyme required for the phosphoryl transfer reaction. Furthermore, the increase of Km with the increase in borate concentration is mediated by the increased formation of the rNTP–borate complex, which reduces TdT′s affinity for rNTP substrates.
Figure 2.
Enzyme kinetics for (A) GTP and (B) dGTP incorporation by TdT in the TB buffer system. The initial rate of GTP and dGTP incorporation was obtained within the range of 0.5–2 min.
Single incorporation of a natural and modified nucleotide into DNA oligomer
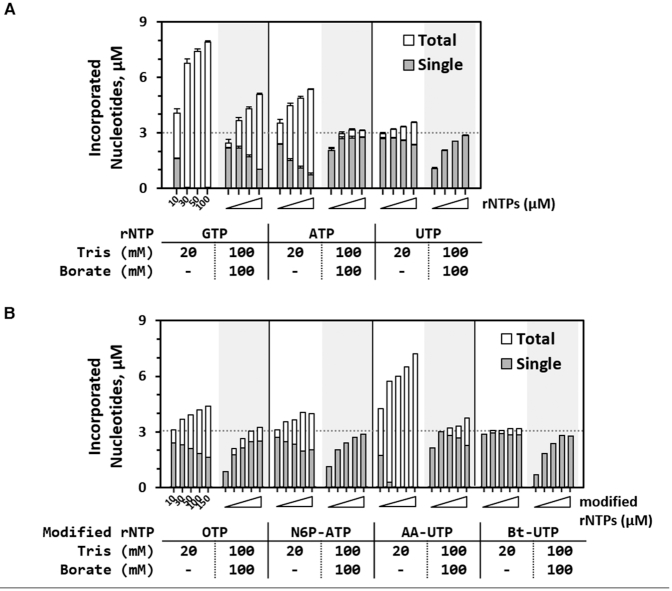
Next, we analyzed 3 kinds of natural rNTPs (Figure 3A) and 4 kinds of modified rNTPs (Figure 3B), which are useful nucleotide units for subsequent applications (for details, see Scheme 2 and Supplementary Figure S3 in Supporting Information). We measured the effects of their borate complex on TdT-catalyzed nucleotide-incorporation. In fact, TdT has been reported to have different rNTP preferences: the order is GTP > ATP > CTP > UTP (12). Here, GTP, ATP, and UTP were evaluated to compare changes in TdT′s preferences for them at different borate concentrations. In addition, their modified rNTPs, such as modified ribonucleotide compounds containing oxanine (OTP, modified GTP), N6-propargyl-adenine (N6P-ATP, modified ATP), aminoallyl-uracil (AA-UTP, modified UTP) or biotin-16-uracil (Bt-UTP, modified UTP) were investigated.
Figure 3.
(A) Quantity of incorporated natural ribonucleotides, and (B) modified ribonucleotides in Tris-borate buffer compared to a conventional reaction buffer for TdT: 20 mM Tris-HCl. Each concentration of total and single nucleotides that are incorporated into ODNs was determined by measuring the relevant peak intensity after PAGE. Because 3 μM initiator (F-10dA, 10 nt) was used, the single incorporated nucleotide that matches the concentration of the initiator depicted as a dotted line means that a high yield of a single-nucleotide incorporation product (11 nt) would be obtained.
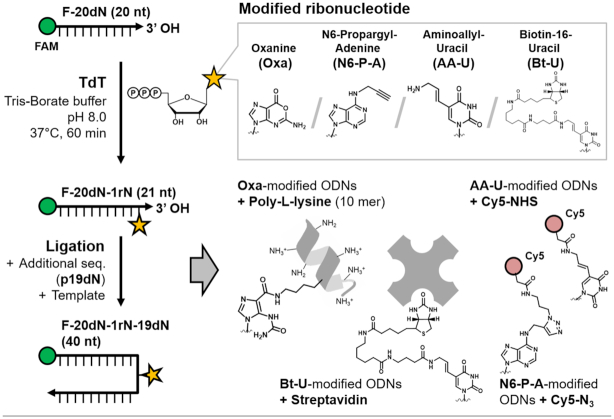
Scheme 2.
Demonstration of single-nucleotide incorporation of commercially available and custom-modified nucleotides via a TdT reaction in TB buffer. Four modified nucleotides useful for bioconjugation were used to label the initiator′s 3′-end, and then placed in the middle of the extended sequence by a ligation method. Each modified ODN was employed for conjugation with its binding or conjugation partner compound.
OTP is prepared by a custom method via nitrosative deamination from GTP (Supplementary Figure S4) (21), and the other three modified rNTPs (N6P-ATP, AA-UTP, and Bt-UTP) are commercially available. Oxanine (Oxa) in OTP is one of the damaged nucleobases arising in vivo and has been intensively studied regarding its biological roles in DNA damage or repair processing (30,33–35). Because Oxa has an O-acylisourea structure, it can react with the molecules containing nucleophilic groups (37). We have reported a chemical synthesis method for preparing Oxa-containing ODNs (21) and proposed the use of Oxa as a novel ODN cross-linker for covalent bonding of ODNs with nucleophilic biomolecules (37), amine-rich nucleoproteins (27,36), or amine-modified solid supporting materials (28).
Each normal or modified nucleotide substrate showed different incorporation efficiency of TdT elongation. In general, the TdT reactions conducted in the conventional 20 mM Tris buffer (without borate ions) showed typical substrate concentration–sensitive incorporation. In particular, when GTP and AA-UTP were used, even a small increase in substrate concentration could result in uncontrollable incessant incorporation of the nucleotides.
In contrast, in the reaction conducted in 100:100 TB buffer (composed of 100 mM Tris and 100 mM borate), the chain elongation by TdT was significantly inhibited after single incorporation of nucleotide, yielding a high single/total ratio, even at high substrate concentrations. That is, both high purity and high yield of single-nucleotide modified ODNs can be obtained. When the incorporation efficiency was compared between a particular normal rNTP and its modified form, a modified rNTP (e.g. GTP versus OTP or ATP versus N6P-ATP), the cases of modified nucleotides yielded a higher single/total ratio. These results may be due to the decreased preference of TdT for modified substrates, modified rNTPs. For example, biotin-16-UTP (Bt-UTP) with a bulky group showed almost single-nucleotide incorporation even without the help of the borate ion.
In addition, we analyzed whether TdT reaction with rNTP in the borate-containing buffer (TdT in rNTP-borate method) can be broadly applicable for ODN modification regardless of the sequence. We checked the efficiency of single-nucleotide incorporation into different ODNs by TdT in rNTP-borate method (Supplementary Figure S5, Supplementary Table S2). We prepared eight different ODN initiators that have been created by random sequence search (Supplementary Table S3); Each initiator has 20 nt in length, no drastic secondary structure (e.g. hairpin or dimer) and ended with one of A, T, G and C that referred at Supplementary Table S3 (A1, A2; T1, T2; G1, G2; C1, C2). For each initiator set, the results of TdT reaction in borate-containing buffer conditions were compared to those in the conventional buffer. Single incorporation efficiency for each nucleotide (G: GTP, O: OTP, P: N6-propargyl-ATP, AA: Aminoallyl-UTP) was analyzed and profiled by quantifying PAGE results. In conventional buffer, the resulting profiles of nucleotide-incorporated ODNs by TdT reaction are varied with low purity (single/total ratio) and some results seem to show sequence-dependent patterns (e.g. the cases of GTP or AA-UTP). In contrast, TdT in rNTP-borate method showed consistent patterns, the generation of single-nucleotide incorporated ODN products with high purity, regardless of the initiator sequences.
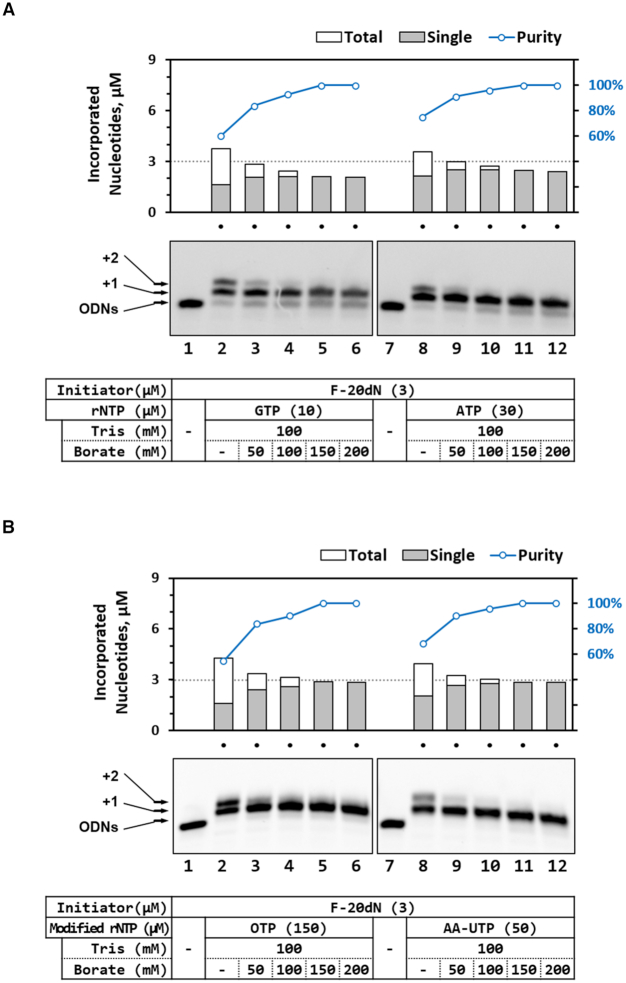
We optimized the concentration of borate to obtain only the single-incorporated ODN product (‘+1’ ODN) via TdT in rNTP-borate method. Although TdT in rNTP-borate method could produce ‘+1’ ODN as a major product, ‘+2’ ODN was also observed (even in trivial amount) for some nucleotides (e.g. for rNTP, GTP and ATP in Figure 1A and 3A; for modified rNTP, OTP and AA-UTP in Figure 3B). Such by-product generation of ‘+2’ ODNs can be successfully restricted by increasing the borate concentration in TdT reaction buffer (Figure 4). That is, as the borate concentration increase, its inhibition effect on TdT is augmented leading to the predominant generation of ‘+1’ ODN. While 100:100 TB buffer is enough for the single-incorporation of UTP, N6P-ATP or Bt-UTP, 100:150 TB buffer is required for the cases of GTP, ATP, OTP or AA-UTP. The optimal conditions (including the borate concentrations) for the rNTP (or modified rNTP) studied here were listed up in Supplementary Table S4, which may be useful guidelines for obtaining only ‘+1’ ODN products.
Figure 4.
Effect of borate concentration on the purity of single-nucleotide incorporation by TdT. Each of rNTP and modified rNTP was inserted to at the 3′-end of F-20dN initiator by TdT reaction in the Tris buffer (20 mM, pH 8.0) containing various concentration of borate ion (0–200 mM). The reaction products were separated on denaturing PAGE in a 12% gel (with 7 M urea) and quantified by analyzing the intensity of FAM dye, which was labeled on the 5′-end initiator. The purity was obtained by measuring the ratio of single- per multi-incorporated ODN product.
These findings implied that the newly developed strategy involving rNTP and the borate ion in a TdT elongation reaction (TdT in rNTP–borate method) can be generally applied to single-nucleotide extension of ODNs, in particular, may be more suitable and effective for incorporation of a modified nucleotide into an ODN in the single-nucleotide mode.
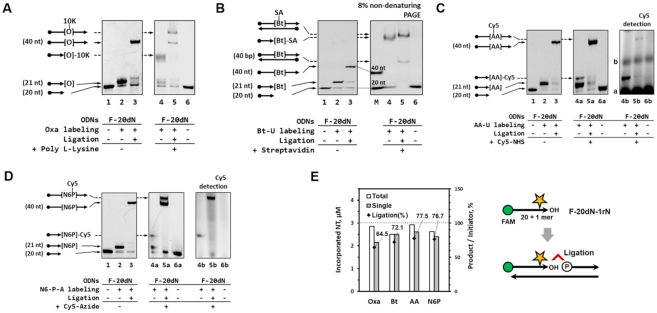
Applications: Preparation of internally modified ODNs and ODN conjugations
We investigated single-nucleotide modified ODNs prepared by the new strategy developed here (TdT in rNTP–borate method), in particular, in terms of their applicability to subsequent biochemical or bioconjugation reactions (Scheme 2). In both fields of biochemical and bioconjugation reactions, it is important to secure modified biomolecules in a quantitatively controlled fashion, which is required for the design of biomolecular systems with desirable performance (25). In particular, for DNA-based systems, it is strongly preferred that available modification methods can insert a single reactive group into a target ODN. To obtain the single-nucleotide incorporated ODN product (21 nt), we employed an optimal concentration of each modified rNTP in a TdT reaction in TB buffer with a 20 nt initiator ODN (for modification). After the TdT reaction in TB buffer, the 21 nt product was purified by means of a centrifugal filter (3 kDa molecular weight cut-off; MWCO), where residual borate and the unreacted rNTP–borate complex were removed. For preparation of internally modified ODNs (a 40 nt ligation product), the purified ‘+1’ ODN product (21 nt) was subsequently ligated with a 19 nt ODN against a 40 nt template ODN via a normal T4 ligase reaction. As a result, the method developed here (TdT in rNTP–borate method) was effective at preparation of the single-nucleotide incorporated ODNs with a modified group, with a >70% yield and almost 100% purity (Lanes 1–3 in Figure 5A–D and E). Because the 21 nt ODN products for 4 kinds of modified rNTPs have an active 3′-hydroxyl group, they were successfully recognized as a ligation fragment, producing a high yield of the extended ODNs that have each modified nucleotide in the middle of the sequence. The ligation results revealed that if borate that is used for single-nucleotide incorporation was removed at the purification step, this approach does not affect further biochemical or enzymatic processes like ligation.
Figure 5.
The TdT reaction in TB buffer for terminal and internal modification of ODNs. To confirm the functionality, (A) oxanine, (B) biotin, (C) aminoallyl or (D) N6-propargyl group–modified ODNs were reacted each with poly-l-lysine, streptavidin, N-hydroxysuccinimide (NHS)-labeled Cy5 dye, or azide-labeled Cy5 dye, respectively. Denaturing PAGE in a 12% gel (with 7 M urea) with detection of FAM dye (excitation 495 nm, emission 520 nm) was mainly used to visualize the reaction products (adducts) ranging from 20 to 40 nt. a Residual NHS-Cy5 dye. b Artifacts present in the NHS-Cy5 solution.
Next, each modified ODN product and its subsequent ligation product was employed for a reaction with its binding or conjugation partner molecule (Figure 5A–D). Oxanine (Oxa) has a unique reactive group, O-acylisourea in the purine ring, so that an Oxa-modified ODN can be conjugated with amine molecules in an activation-free fashion (26–31). When Oxa-modified ODNs prepared here were incubated with poly-l-lysine (10 mer) for 48 h, the conjugate formation was observed as the single band with increased size (lanes 4–6 in Figure 5A). Biotin (Bt) can strongly and specifically bind to streptavidin. Because this conjugation is based on a noncovalent interaction, the conjugation products were analyzed by native PAGE. As expected, Bt-U-modified ODNs were successfully observed as the conjugate complex with streptavidin (∼53 kDa; lanes 4–6 in Figure 5B). It is known that aminoallyl group of AA-UTP can react with the N-hydroxysuccinimide (NHS) group. When the modified ODNs with single AA-U were incubated with the NHS-Cy5 dye, a single band of the Cy5-labeled product was observed. The band was confirmed by the FAM fluorescence signal of ODNs and the Cy5 fluorescence signal of the conjugate (lanes 4–6 in Figure 5C). In case of N6P-A incorporation, the propargyl group in the prepared ODNs can react with an azide group in the CuAAC method (click chemistry) (32). When the N6P-A–modified ODNs were incubated with the azide-Cy5 dye, the Cy5-labeled ODN conjugate was observed and confirmed by both FAM and Cy5 fluorescence signal analysis (lanes 4–6 in Figure 5D).
DISCUSSION
The enzymatic modification of DNA oligomers, in particular, DNA extension or replacements by DNA polymerases have been useful ways for the post-synthetic design of ODNs. Among the polymerases, terminal deoxynucleotidyl transferase (TdT) is frequently used since it can catalyze chain elongation of DNA fragments easily without a template by adding random nucleotides at the 3′-end of the strand. Low substrate specificity and template independence of TdT are the merits for the simple extension of DNA, but TdT cannot be used for the specific modification of ODNs. Until now, the applications of TdT have been limited to the use of its random chain elongation, such as signal amplification (8,9) and polymerization of building blocks (10,11).
In a previous work, we showed a possibility to use TdT for modification of ODN in more controlled or designed ways. We prepared a reactive-end DNA oligomer by optimizing TdT reaction with deoxyribonucleotide substrate of Oxa (dOTP), where single Oxa-incorporated ODN was observed as a main product (31). However, there were still limitations for achieving single-nucleotide incorporation (‘+1’ ODN product) using TdT. Basically, the traits of TdT leading to incessant incorporation of nucleotides cannot be overcome by just optimizing the reaction conditions. Moreover, the design strategy was not generally applicable for other modified nucleotides. That was just specialized for Oxa-modified ODN based on the unique property (substrate inhibition) of dOTP for TdT. More robust and general method for single-nucleotide modification of ODN by TdT has not yet been developed.
Herein, we present a novel enzymatic method for single-nucleotide incorporation of ODNs with a modified (or normal) nucleotide; this approach is useful for biochemical or bioconjugation applications. The newly developed method features the use of an rNTP substrate and a borate-containing buffer in a TdT elongation reaction. The use of rNTPs as substrates instead of dNTPs in a TdT reaction could reduce uncontrolled chain incorporation of nucleotides (lanes 5 versus 10 of Figure 1A). This is that TdT has a lower preference for rNTPs than dNTPs (12,13). In high-concentrated Tris buffer, slight inhibition was also observed (lanes 4 versus 5 of Figure 1A), as reported in other researches (23,24), but this inhibition is not enough for achieving the single-nucleotide incorporation. We hypothesized that formation of the rNTP–borate complex would more effectively inhibit the chain nucleotide incorporation reaction by TdT. As general, ribonucleotide–borate complex via a reversible borate ester with cis-2′,3′-diol can contribute to the formation of stable RNA structure (15–20), which is maintained even at relatively low pH of 6.6 and high temperature (60°C) in saline (19,20). It was found for the first time here that the complex of a ribonucleotide substrate (rNTP) with the borate ion (Scheme 1A) can prevent incessant nucleotide incorporation in a chain elongation reaction of TdT (lanes 2–3 of Figure 1A, Figure 1B; Figure 2 and 3A). The process was simply implemented by carrying out a TdT reaction with rNTP in TB buffer (Scheme 1B).
Moreover, rNTP with various modified groups [modified rNTP; N6P-ATP, AA-UTP and Bt-UTP (commercially available) or OTP (custom made)] can be applied to the design of single-nucleotide modified ODNs. The chain elongation by TdT was significantly inhibited, and the single-nucleotide incorporated ODN was achieved reliably. Of note, a higher single/total ratio was observed for a modified form, a modified rNTP than its normal rNTP (e.g. OTP versus GTP or N6P-ATP versus ATP in Figure 3). It might be due to the decreased preference of TdT for modified substrates (modified rNTPs) [e.g. the low preference of TdT for Bt-UTP (with a bulky group) leading to high efficiency (even without borate ion) in Figure 3]. It was also checked whether TdT in rNTP-borate method can be generally applicable for other ODN modifications regardless of the sequence. Eight different ODN initiators (with no drastic secondary structure) that were designed by random sequence search were tested (Supplementary Table S3). Single-nucleotide incorporated ODNs can be obtained by TdT in rNTP-borate method, regardless of the initiator sequences (Supplementary Figure S5).
The reaction conditions for TdT in rNTP-borate method were optimized to obtain single-nucleotide modified DNA oligomers (‘+1’ ODN products) with a high purity. Some by-product generation of ‘+2’ or ‘+3’ ODNs can be successfully restricted by increasing the borate concentration in TdT reaction buffer (Figure 4). This is the first results to use TdT for single-nucleotide modification of ODN in more robust and reliable way. The optimal conditions (including the borate concentration and substrate concentration) for the rNTP (or modified rNTP) studied here were summarized in Supplementary Table S4.
Moreover, because the borate ester bonding in the complex is reversible, the single-nucleotide modified ODNs, after removal of borate by purification, can be employed in further applications such as enzymatic reaction or biophysical/chemical conjugation. The single-incorporated ODN products (21 nt) with 4 kinds of modified rNTPs have been successfully recognized by ligase enzyme as a ligation fragment, producing a high yield of the extended ODNs that have each modified nucleotide in the middle of the sequence. In addition, each nucleotide-modified products or its subsequent ligation products have been employed successfully for a subsequent reaction with its binding or conjugation partner compound (Oxa-modified ODNs with poly-l-lysine, Bt-U-modified ODNs with streptavidin, AA-U-modified ODNs with the NHS-Cy5 dye, and N6P-A-modified ODNs with azide-Cy5 dye) (Scheme 2, Figure 5A–D).
The results obtained here indicate that the novel ‘TdT in rNTP-borate’ method can be quite simple and efficient enzymatic strategy for designing a single-nucleotide modified DNA oligomer, which can subsequently be used in one-to-one bioconjugation applications. Such a single-nucleotide modified ODN is useful for the biophysical/biochemical studies or the development of a quantitative and reproducible DNA-conjugated system.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT [NRF-2016R1A2B4014237]; Basic Core Technology Development Program for the Oceans and the Polar Regions of the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT, Republic of Korea [NRF-2015M1A5A1037054]; BK21 plus. Funding for open access charge: Korea University Grant.
Conflict of interest statement. None declared.
REFERENCES
- 1. Rajendran A., Endo M., Sugiyama H.. Single-Molecule analysis using DNA origami. Angew. Chem. Int. Ed. 2012; 51:874–890. [DOI] [PubMed] [Google Scholar]
- 2. Silverman S.K. Pursuing DNA catalysts for protein modification. Acc. Chem. Res. 2015; 48:1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uzawa T., Tada S., Wang W., Ito Y.. Expansion of the aptamer library from a “natural soup” to an “unnatural soup”. Chem. Commun. 2013; 49:1786–1795. [DOI] [PubMed] [Google Scholar]
- 4. Ni S.J., Yao H.Z., Wang L.L., Lu J., Jiang F., Lu A.P., Zhang G.. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int. J. Mol. Sci. 2017; 18:1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gilfillan S., Dierich A., Lemeur M., Benoist C., Mathis D.. Mice lacking Tdt - mature animals with an immature lymphocyte repertoire. Science. 1993; 261:1175–1178. [DOI] [PubMed] [Google Scholar]
- 6. Motea E.A., Berdis A.J.. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. BBA-Proteins Proteom. 2010; 1804:1151–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fowler J.D., Suo Z.. Biochemical, structural, and physiological characterization of terminal deoxynucleotidyl transferase. Chem. Rev. 2006; 106:2092–2110. [DOI] [PubMed] [Google Scholar]
- 8. Horakova P., Macickova-Cahova H., Pivonkova H., Spacek J., Havran L., Hocek M., Fojta M.. Tail-labelling of DNA probes using modified deoxynucleotide triphosphates and terminal deoxynucleotidyl tranferase. Application in electrochemical DNA hybridization and protein-DNA binding assays. Org. Biomol. Chem. 2011; 9:1366–1371. [DOI] [PubMed] [Google Scholar]
- 9. Peng F.F., Liu Z.L., Li W., Huang Y., Nie Z., Yao S.Z.. Enzymatically generated long polyT-templated copper nanoparticles for versatile biosensing assay of DNA-related enzyme activity. Anal. Methods-UK. 2015; 7:4355–4361. [Google Scholar]
- 10. Hu W.W., Ning Y., Kong J.M., Zhang X.J.. Formation of copper nanoparticles on poly(thymine) through surface-initiated enzymatic polymerization and its application for DNA detection. Analyst. 2015; 140:5678–5684. [DOI] [PubMed] [Google Scholar]
- 11. Tang L., Navarro L.A., Chilkoti A., Zauscher S.. High-Molecular-Weight polynucleotides by Transferase-Catalyzed living Chain-Growth polycondensation. Angew. Chem. Int. Ed. 2017; 56:6778–6782. [DOI] [PubMed] [Google Scholar]
- 12. Boule J.B., Rougeon F., Papanicolaou C.. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem. 2001; 276:31388–31393. [DOI] [PubMed] [Google Scholar]
- 13. Winz M.L., Linder E.C., Andre T., Becker J., Jaschke A.. Nucleotidyl transferase assisted DNA labeling with different click chemistries. Nucleic Acids Res. 2015; 43:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuo J., Murakami M.. The mukaiyama aldol Reaction: 40Years of continuous development. Angew. Chem. Int. Ed. 2013; 52:9109–9118. [DOI] [PubMed] [Google Scholar]
- 15. Martin A.R., Vasseur J.J., Smietana M.. Boron and nucleic acid chemistries: merging the best of both worlds. Chem. Soc. Rev. 2013; 42:5684–5713. [DOI] [PubMed] [Google Scholar]
- 16. Cossetti C., Crestini C., Saladino R., Di Mauro E.. Borate minerals and RNA stability. Polymers-Basel. 2010; 2:211–228. [Google Scholar]
- 17. Ricardo A., Carrigan M.A., Olcott A.N., Benner S.A.. Borate minerals stabilize ribose. Science. 2004; 303:196–196. [DOI] [PubMed] [Google Scholar]
- 18. Scorei R., Cimpoiasu V.M.. Boron enhances the thermostability of carbohydrates. Origins Life Evol. B. 2006; 36:1–11. [DOI] [PubMed] [Google Scholar]
- 19. Amaral A.F., Marques M.M., da Silva J.A.L., da Silva J.J.R.F.. Interactions of D-ribose with polyatomic anions, and alkaline and alkaline-earth cations: possible clues to environmental synthesis conditions in the pre-RNA world. New J. Chem. 2008; 32:2043–2049. [Google Scholar]
- 20. Furukawa Y., Horiuchi M., Kakegawa T.. Selective stabilization of ribose by Borate. Origins Life Evol.Biospheres. 2013; 43:353–361. [DOI] [PubMed] [Google Scholar]
- 21. Pack S.P., Nonogawa M., Kodaki T., Makino K.. Chemical synthesis and thermodynamic characterization of oxanine-containing oligodeoxynucleotides. Nucleic Acids Res. 2005; 33:5771–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suzuki T., Yoshida M., Yamada M., Ide H., Kobayashi M., Kanaori K., Tajima K., Makino K.. Misincorporation of 2 ‘-deoxyoxanosine 5 ’-triphosphate by DNA polymerases and its implication for mutagenesis. Biochemistry-US. 1998; 37:11592–11598. [DOI] [PubMed] [Google Scholar]
- 23. Desmarais W.T., Bienvenue D.L., Bzymek K.P., Holz R.C., Petsko G.A., Ringe D.. The 1.20 angstrom resolution crystal structure of the aminopeptidase from Aeromonas proteolytica complexed with tris: a tale of buffer inhibition. Structure. 2002; 10:1063–1072. [DOI] [PubMed] [Google Scholar]
- 24. Ghalanbor Z., Ghaemi N., Marashi S.A., Amanlou M., Habibi-Rezaei M., Khajeh K., Ranjbar B.. Binding of Tris to Bacillus licheniformis alpha-amylase can affect its starch hydrolysis activity. Protein Peptide Lett. 2008; 15:212–214. [DOI] [PubMed] [Google Scholar]
- 25. Kalia J., Raines R.T.. Advances in bioconjugation. Curr. Org. Chem. 2010; 14:138–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kamisetty N.K., Pack S.P., Nonogawa M., Yamada K., Yoshida Y., Kodaki T., Makino K.. Stabilization of the immobilized linkers and DNA probes for DNA microarray fabrication by end-capping of the remaining unreacted silanol on the glass. J. Biotechnol. 2009; 140:242–245. [DOI] [PubMed] [Google Scholar]
- 27. Nakano T., Terato H., Asagoshi K., Masaoka A., Mukuta M., Ohyama Y., Suzuki T., Makino K., Ide H.. DNA-protein cross-link formation mediated by oxanine - a novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem. 2003; 278:25264–25272. [DOI] [PubMed] [Google Scholar]
- 28. Pack S.P., Kamisetty N.K., Nonogawa M., Devarayapalli K.C., Ohtani K., Yamada K., Yoshida Y., Kodaki T., Makino K.. Direct immobilization of DNA oligomers onto the amine-functionalized glass surface for DNA microarray fabrication through the activation-free reaction of oxanine. Nucleic Acids Res. 2007; 35:e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pack S.P., Morimoto H., Makino K., Tajima K., Kanaori K.. Solution structure and stability of the DNA undecamer duplexes containing oxanine mismatch. Nucleic Acids Res. 2011; 40:1841–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pack S.P., Doi A., Choi Y.S., Kodaki T., Makino K.. Biomolecular response of oxanine in DNA strands to T4 polynucleotide kinase, T4 DNA ligase, and restriction enzymes. Biochem. Biophys. Res. Commun. 2010; 391:118–122. [DOI] [PubMed] [Google Scholar]
- 31. Jang E.K., Ki M.-R., Pack S.P.. Design of reactive-end DNA oligomers via incorporation of oxanine into oligonucleotides using terminal deoxynucleotidyl transferase. Process Biochem. 2017; 62:99–105. [Google Scholar]
- 32. Liang L.Y., Astruc D.. The copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) “click” reaction and its applications: an overview. Coord. Chem. Rev. 2011; 255:2933–2945. [Google Scholar]
- 33. Suzuki T., Yamaoka R., Nishi M., Ide H., Makino K.. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxynucleotide, and calf thymus DNA treated by nitrous acid and nitric oxide. J. Am. Chem. Soc. 1996; 118:2515–2516. [Google Scholar]
- 34. Nakano T., Ouchi R., Kawazoe J., Pack S.P., Makino K., Ide H.. T7 RNA polymerases backed up by covalently trapped proteins catalyze highly error prone transcription. J. Biol. Chem. 2012; 287:6562–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nakano T., Miyamoto-Matsubara M., Shoulkamy M.I., Salem A.M.H., Pack S.P., Ishimi Y., Ide H.. Translocation and stability of replicative DNA helicases upon encountering DNA-protein cross-links. J. Biol. Chem. 2013; 288:4649–4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakano T., Morishita S., Terato H., Pack S.P., Makino K., Ide H.. Repair mechanism of DNA-protein cross-link damage in Escherichia coli. Nucleic Acids Symp. Ser. 2007; 51:213–214. [DOI] [PubMed] [Google Scholar]
- 37. Pack S.P., Doi A., Kamisetty N.K., Nonogawa M., Kodaki T., Makino K.. Functional reactivity of oxanine: its biological meanings and biotechnological applications. Nucleic Acids Symp. Ser. 2007; 51:53–54. [DOI] [PubMed] [Google Scholar]
- 38. Gozalbo-López B., Andrade P., Terrados G., de Andrés B., Serrano N., Cortegano I., … & Gaspar M. L.. A role for DNA polymerase μ in the emerging DJH rearrangements of the postgastrulation mouse embryo. Mol. Cell Biol. 2009; 29:1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.