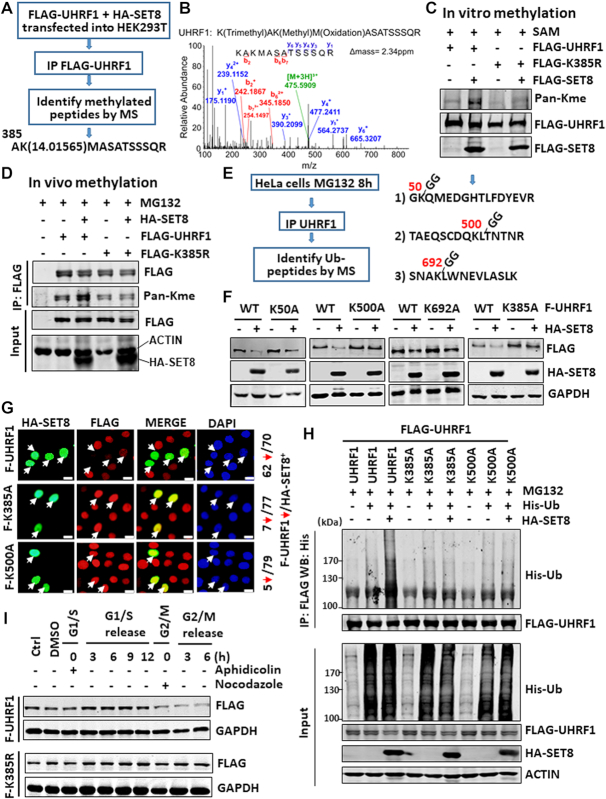
Figure 3.
Identification and characterization of UHRF1 methylation and ubiquitination sites. (A) Strategy for identification of UHRF1 methylation site(s). Note that a peptide with monomethylated K385 was identified from FLAG-UHRF1 co-expressed with SET8. (B) Mass spectrometry profile of the K385me1-containing peptide. (C) In vitro methylation assay showing that SET8 methylated UHRF1 in a K385 site-dependent manner. FLAG-SET8, FLAG-UHRF1 or FLAG-UHRF1-K385R mutant was individually expressed in HEK293T cells, purified by anti-FLAG M2 beads and subjected to in vitro methylation assay. (D) SET8 enhanced UHRF1 methylation in vivo in a K385-dependent manner. Wild-type or UHRF1 K385R mutant was co-expressed with or without SET8 in HEK283T cells and the methylation on UHRF1 was detected by IP-western blot analysis. (E) Strategy and summary of UHRF1 ubiquitination sites identified by mass spectrometry. (F) Western blot analysis showing that K385A and K500A mutants were resistant to SET8-induced UHRF1 degradation. HeLa cells stably expressing FLAG-UHRF1 or various mutants were established first and then transfected with a SET8-expressing plasmid. (G) Immunostaining showing that ectopically expressed HA-SET8 downregulated wild-type FLAG-UHRF1 but not K385A and K500A mutants in the corresponding stable cell lines. (H) Ubiquitination assay showing that K385A and K500A mutants were resistant to SET8-induced ubiquitination of UHRF1. (I) Western blot analysis showing that the K385R mutant was not downregulated in G2/M phase.