Table 1.
Summary of unsupervised methods
| Definition of Genomic Regions | Test Association Between Methylations in Genomic Regions vs. Continuous Phenotypea | Reference | |
|---|---|---|---|
| Previously proposed methods | |||
| IMA_mean (rforge.net/IMA/) | Illumnina annotation (e.g. CGI, TSS200) | linear model: 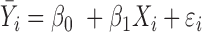 where where 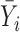 is mean of methylation levels over all CpGs in the region, for sample is mean of methylation levels over all CpGs in the region, for sample 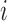
|
(26) |
| IMA_median (rforge.net/IMA/) | Illumnina annotation (e.g. CGI, TSS200) | linear model: 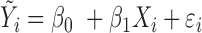 where where 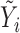 is median of methylation levels over all CpGs in the region, for sample is median of methylation levels over all CpGs in the region, for sample 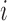
|
(26) |
| Aclust (github.com/tamartsi/Aclust/) | Adjacent Site Clustering (A-clustering) algorithm | Generalized Estimating Equation model: 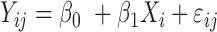 , where , where 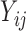 = methylation value for CpG = methylation value for CpG 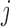 in sample in sample 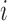 ; ; 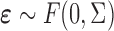 for some mean-zero distribution F with covariance matrix for some mean-zero distribution F with covariance matrix 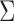 . . |
(20) |
| Seqlm (github.com/raivokolde/seqlm) | Minimum Description Length principle | simple linear mixed model: 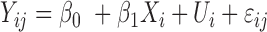 , where , where 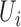 is the sample random effect is the sample random effect |
(19) |
| Proposed in this study | |||
| coMethDMR_simple (github.com/lissettegomez/coMethDMR) | CoMethAllRegions function | simple linear mixed model: 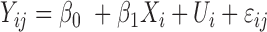 , where , where 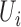 is the sample random effect is the sample random effect |
|
| coMethDMR_randCoef (github.com/lissettegomez/coMethDMR) | CoMethAllRegions function | random coefficient mixed model: 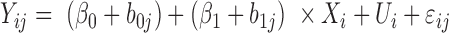 where where 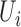 is the sample random effect; is the sample random effect; 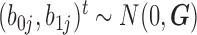 are random coefficients for intercepts and slopes; are random coefficients for intercepts and slopes; 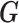 is an unstructured covariance matrix. is an unstructured covariance matrix. |
a
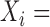 continuous phenotype (e.g. disease stage) for sample
continuous phenotype (e.g. disease stage) for sample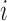 ;
;
