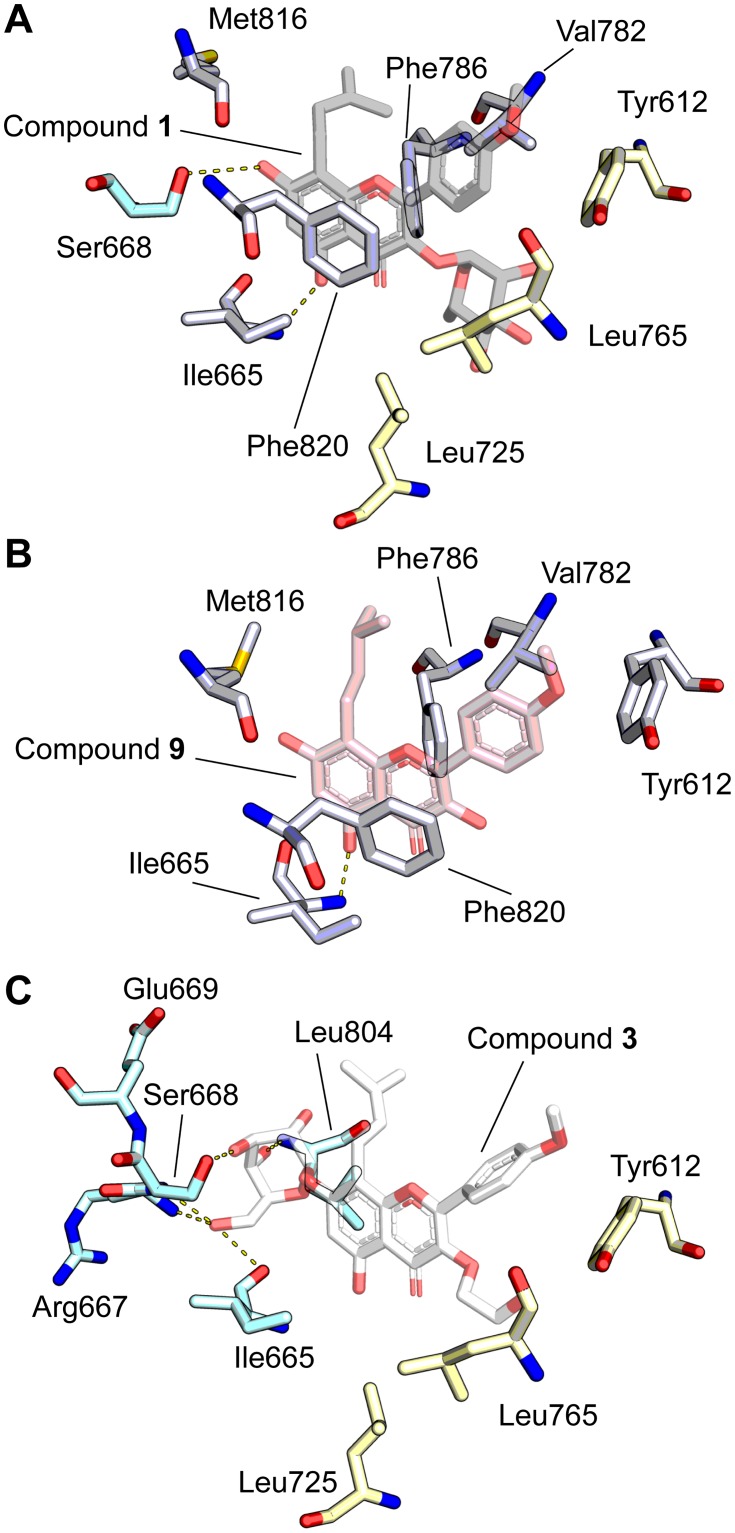
Fig 4. Molecular modeling of PDE5 with icariin analogs using PDB ID: 2H44 [20] as the starting model.
(A) Crystal structure of compound 1 (grey) bound to PDE5 (PDB ID: 2H44) [20], highlighting ligand orientation and nearby interacting residues. Residues shared with panel B are white while residues shared with panel C are blue (interacts with 7-O position) and yellow (hydrophobic pocket; interacts with 3-O position). (B) Ligand docking model of compound 9 (pink) bound to PDE5 illustrates residues (white) that interact with conserved elements of the icariin-derived compounds, such as the flavone backbone and prenyl group. (C) Ligand docking model of compound 3 (white) bound to PDE5 illustrates residues that form hydrogen bonds with the 7-O-glucose (blue) and that form a hydrophobic pocket surrounding the 3-O-hydroxyethyl group (yellow). Hydrogen bonding is indicated with dashed yellow lines.