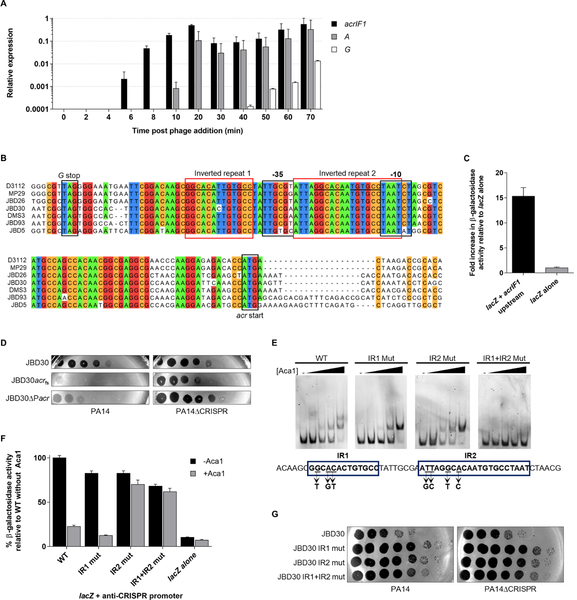
Figure 2. acrIF1 expression is driven by a promoter region that includes binding sites for Aca1 (A).
Relative transcription levels of phage genes were measured by RT-qPCR at the indicated times after infection of PA14 by phage JBD30. Transcriptional levels are shown of the anti-CRISPR gene (acrIF1), an early expressed gene (A, transposase), and a late expressed gene (G, a tail component) during one round of phage infection at a multiplicity of infection (MOI) of 5. Levels were normalized to the geometric mean of the transcript levels of the housekeeping genes: clpX and rpoD. Data are represented as mean ± SEM from three independent experiments. (B) Multiple nucleotide sequence alignment of anti-CRISPR phages from the stop codon of the Mu G homologue (G stop) to the start codon of the anti-CRISPR genes (acr start). Bioinformatically predicted promoter elements (BPROM; Solovyev and Salamov, 2011) −10 and −35 are shown. Inverted repeats are indicated by red boxes. A common sequence in both repeats is underlined. Positions sharing greater than 85% identity are coloured according to nucleotide. (C) The putative acr promoter region from phage JBD30 was cloned upstream of a promoterless lacZ expression vector (lacZ+acrIF1 upstream) and β-galactosidase activity was measured in P. aeruginosa strain PA14. The mean ± SEM of three independent assays is shown. (D) Tenfold dilutions of wild-type (JBD30), anti-CRISPR gene frameshift mutant (JBD30acrfs) and acr promoter mutant (JBD30ΔPacr) phage lysates were applied to lawns of PA14 and PA14ΔCRISPR. (E) EMSAs were performed using a dsDNA fragment with the sequence shown, which encompasses the acr promoter region. The IR1 and IR2 mutants contained the triple and quadruple base substitutions indicated under the DNA sequence. Representative non-denaturing polyacrylamide gels stained with SYBR gold are shown. Purified Aca1 was added to the DNA at concentrations of 10 nM, 50 nM, 100 nM and 250 nM. The dash sign (−) indicates no added protein. (F) The acr promoter region from JBD30 either wild-type (WT), or bearing IR1 and/or IR2 mutations was cloned upstream of a promoterless lacZ gene. β-galactosidase activity was measured in PA14 (−Aca1) or in a JBD30 lysogen (+Aca1). The mean ± SEM β-galactosidase activity relative to the wild-type promoter is shown (n ≥ 3). (G) Tenfold dilutions of phage JBD30 lysates carrying the indicated inverted repeat mutations were applied to lawns of PA14 or PA14ΔCRISPR. Representative images of three replicates are shown. See also Figures S1and S2.