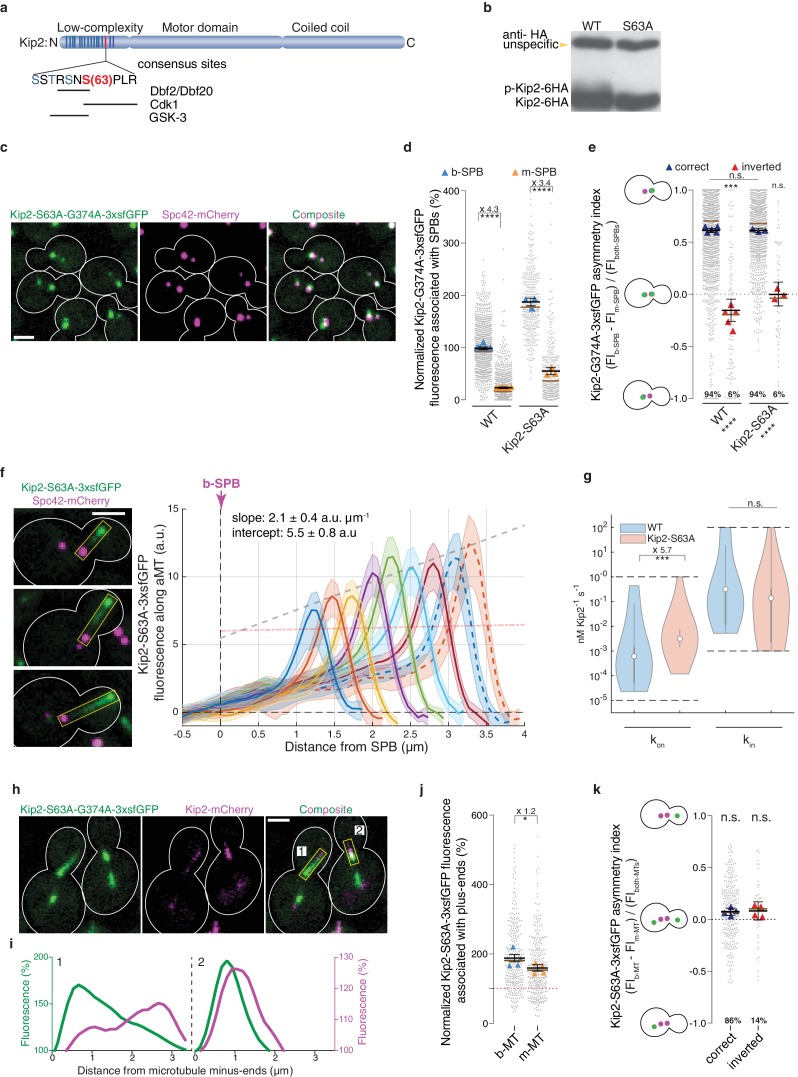
Figure 6. Phosphorylation of its N-terminus prevents Kip2 from landing along microtubules.
(a) Scheme of Kip2 protein and its disordered N-terminus with residue serine 63 (red) and kinase consensus sites (blue). (b) Western blot analysis of endogenously expressed Kip2-6HA and Kip2-S63A-6HA. Lysates were prepared from cycling cells of indicated genotype. (c) Representative images of Kip2-S63A-G374A-3xsfGFP (green) in preanaphase cells. Spindles are visualized with Spc42-mCherry (magenta). See Figure 3b for control. (d) Normalized Kip2-S63A-G374A-3xsfGFP fluorescence (%) associated with b- (blue) and m-SPBs (yellow) in cells shown in (c) (n > 3 independent clones or technical replicates,>314 cells per genotype). (e) Quantification of asymmetry index for Kip2-S63A-G374A-3xsfGFP distribution (fluorescence intensity: (FIb-SPB – FIm-SPB) / FIboth-SPBs) between SPBs for data in (d) with correctly orientated (blue) and inverted (red) SPBs. (f) Representative images (left) and quantifications (right) of fluorescence intensities (a.u.) from endogenous Kip2-S63A-3xsfGFP along preanaphase aMTs (boxed areas). The graph format is the same as those in Figure 1. 29 ≤ n ≤ 55 per bin. The pink dashed line denotes the weighted linear regression for the mean GFP fluorescence on plus-ends in wild-type cells. See Figure 5—figure supplement 1 for more details. (g) In silico likelihood of on rate constant and in rate constant estimated from model fits to Kip2-3xsfGFP data in wt and Kip2-S63A-3xsfGFP cells. Statistical significance for (***, p<2∙10−4) was determined by sampling from the likelihood for (see Appendix 1), difference in median as indicated. Graph as in Figure 2d. (h) Representative images of preanaphase heterozygous diploid cells expressing the endogenous ATPase deficient protein Kip2-S63A-G374A-3xsfGFP (green) and the wild type protein Kip2-mCherry (magenta). (i) Line scan analysis along the numbered microtubules shown in (h). As in Figure 3b, GFP (green) and mCherry (magenta) fluorescence intensities were normalized to their background levels, respectively. (j) Normalized Kip2-S63A-3xsfGFP fluorescence (%) associated with microtubule plus-ends in preanaphase cells carrying both b- (blue) and m- microtubules (yellow) (305 cells from n > 3 independent clones). Values were normalized to Kip2-3xsfGFP fluorescence associated with b-microtubules in wild-type cells, denoted as red dotted line in the graph. See Figure 5—figure supplement 1a for representative images. (k) Quantification of asymmetry index for Kip2-S63A-3xsfGFP accumulation on microtubule plus-ends (fluorescence intensity: (FIb-MT – FIm-MT) / FIboth-MTs) for data in (j) with correctly orientated (blue) and inverted (red) SPBs. Graphs and statistical analysis are like those in Figure 4 (b,c,e,f,g and i). Statistical significances within each genotype are marked at the bottom of graphs. Source data for these panels are available in Supplementary file 1. Scale bars, 2 µm. See also Video 3, Figure 5—figure supplement 1 and Figure 6—figure supplement 1.