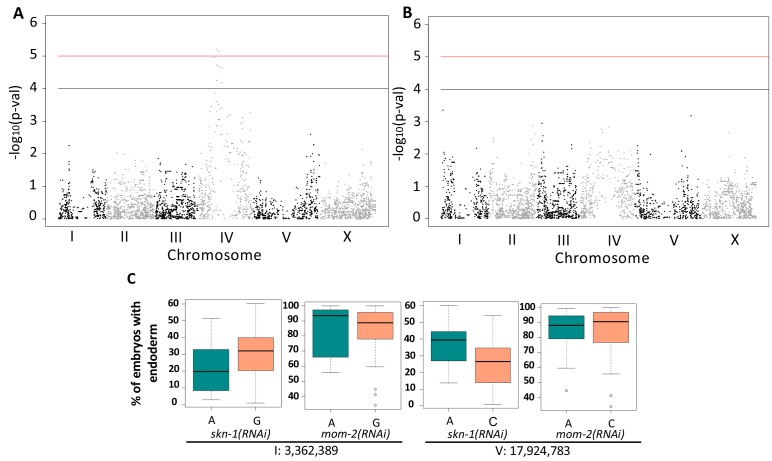
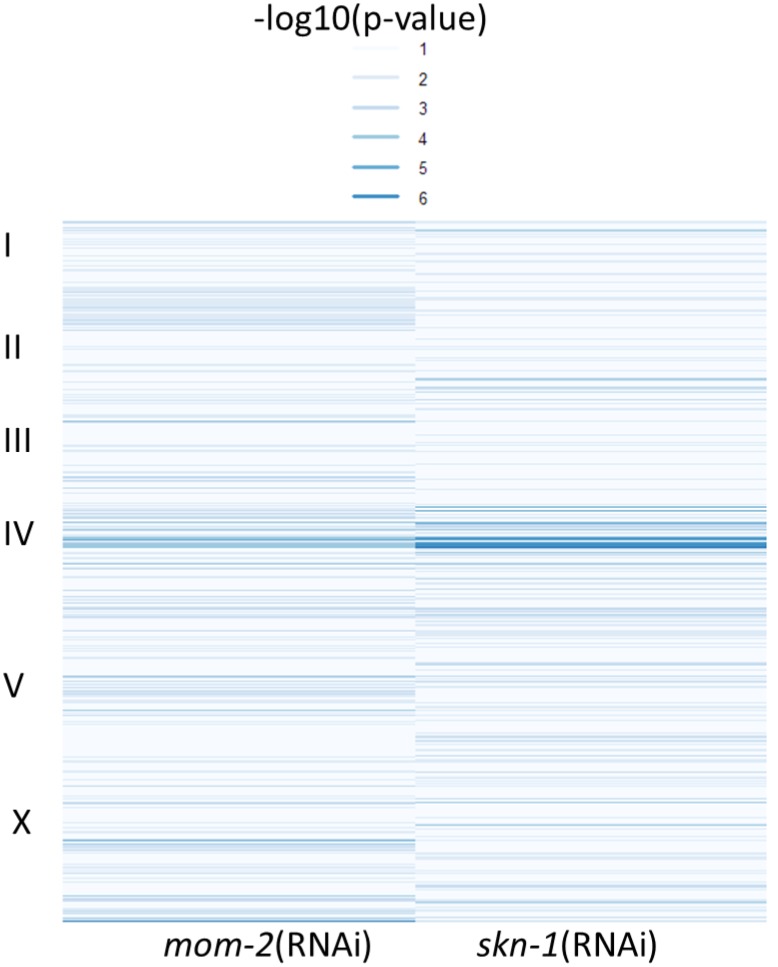
Figure 5. Genome-Wide Association Studies of skn-1(RNAi) and mom-2(RNAi) phenotypes.
(A) Manhattan plot of skn-1(RNAi) GWAS. The red line indicates a genome-wide 1.5% FDR (permutation-based FDR, from 10,000 permutated results). Black line represents 3.0% FDR. The y axis is the –log10 of p-value. (B) Manhattan plot of mom-2 (RNAi) EMMA. The y axis is the –log10 of p-value. Genomic regions are shown on the x-axis. (C) Effect plots of the most strongly-linked SNPs from mom-2(RNAi) GWAS at position 3,362,389 bp on chromosome I and position 17,924,783 bp on chromosome V. Horizontal lines within each box represent the median, and the boxes represent 25th–75th percentile.