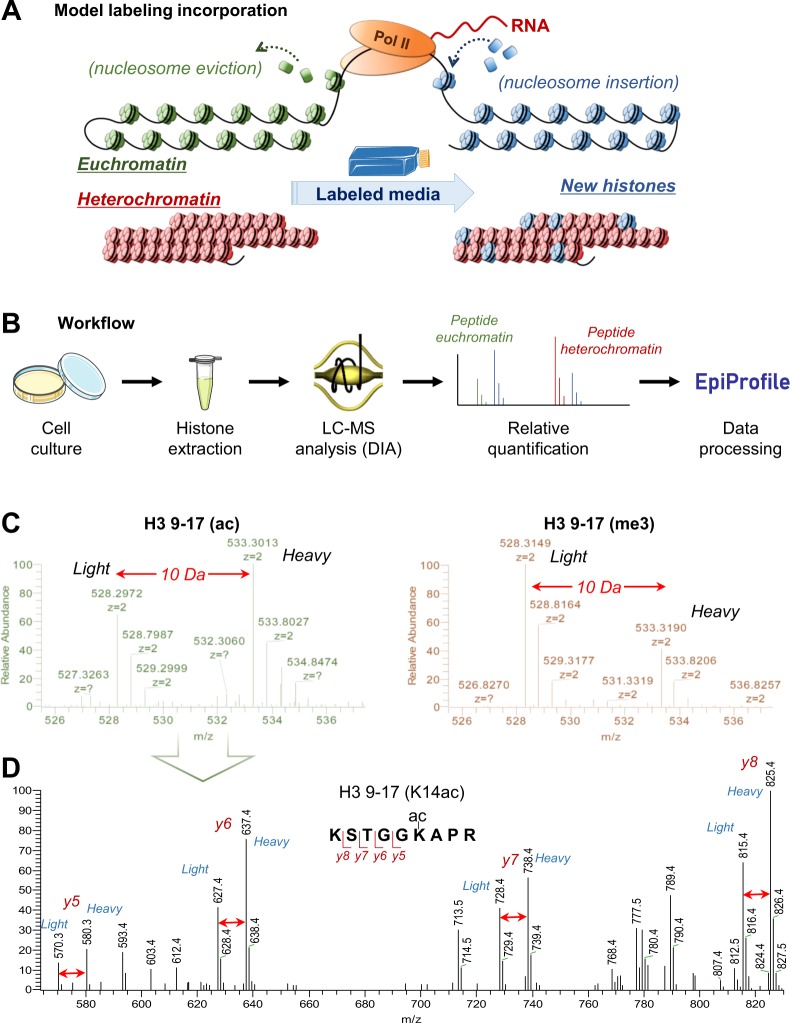
Figure 1.
Overview of the workflow and representative result spectra. (A) Rationale of the metabolic labeling principle; accessible chromatin has higher nucleosome turnover and thus it is labeled with higher rate as compared to silenced heterochromatin. Labeling is represented with the blue color. (B) Schematic of the workflow. Cells are grown in culture for a relatively short amount of time (1–3 days) with selected isotopically heavy amino acids (in this work, arginine) and then processed by using our in-house workflow for canonical histone PTM analysis. Relative quantification (relative PTM abundance and heavy/light ratio) is performed automatically by our in-house software EpiProfile30. (C) Full MS spectrum of the peptide of histone H3 KSTGGKAPR (aa 9–17) carrying one acetyl group (on the left) or one trimethyl group (on the right). Evidently, the peptide in its trimethylated form has a lower % heavy arginine incorporation. (D) MS/MS spectrum of the same peptide carrying one acetyl group, co-isolating the light and the heavy form. The fragment ions can be used to confirm the relative amount of labeling even in presence of isobaric forms (K9ac vs K14ac). Panel A and B were produced, in part, by using Servier Medical Art (http://smart.servier.com/). Servier Medical Art is licensed under a Creative Commons Attribution 3.0 License (CC BY 3.0 license: https://creativecommons.org/licenses/by/3.0/). The color of the images downloaded was modified to fit our figure.