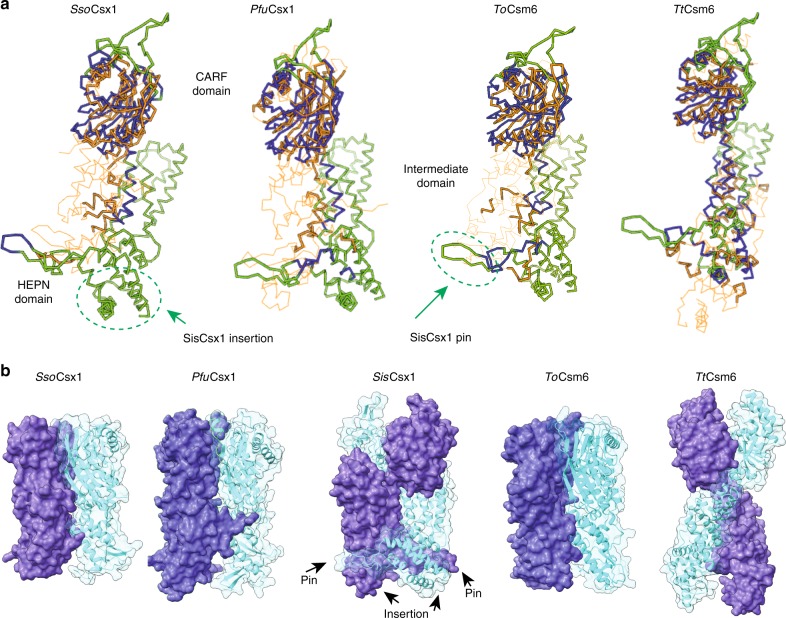
Fig. 2.
Comparison between type III CRISPR-associated ribonucleases. a Structural comparison between SisCsx1 and the dimeric SsoCsx1, PfuCsx1, ToCsm6, TtCsm6. Ribbon representation of the SisCsx1 (green) and PfuCsx1, SsoCsx1 and TtCsm6 (orange) monomers which have been superposed using the DALI server. SsoCsx1 (PDB 2I71) r.m.s.d. to SisCsx1 is 5.1 Å for 184 residues out of 376 with an identity of 20%, for PfuCsx1 (PDB 4EOG) r.m.s.d. to SisCsx1 is 6.1 Å for 200 residues out of the 466 of the protein with an identity of 12%, for ToCsm6 (PDB 6O6S) r.m.s.d. to SisCsx1 is 6.0 Å for 194 residues out of the 433 of the protein with an identity of 15%, while for TtCsm6 (PDB 5FSH) the r.m.s.d. is 5.0 Å for 224 out of the 448 of the protein with an identity of 11%. b Comparison of the dimeric arrangements of SsoCsx1, PfuCsx1, ToCsm6, TtCsm6 with SisCsx1