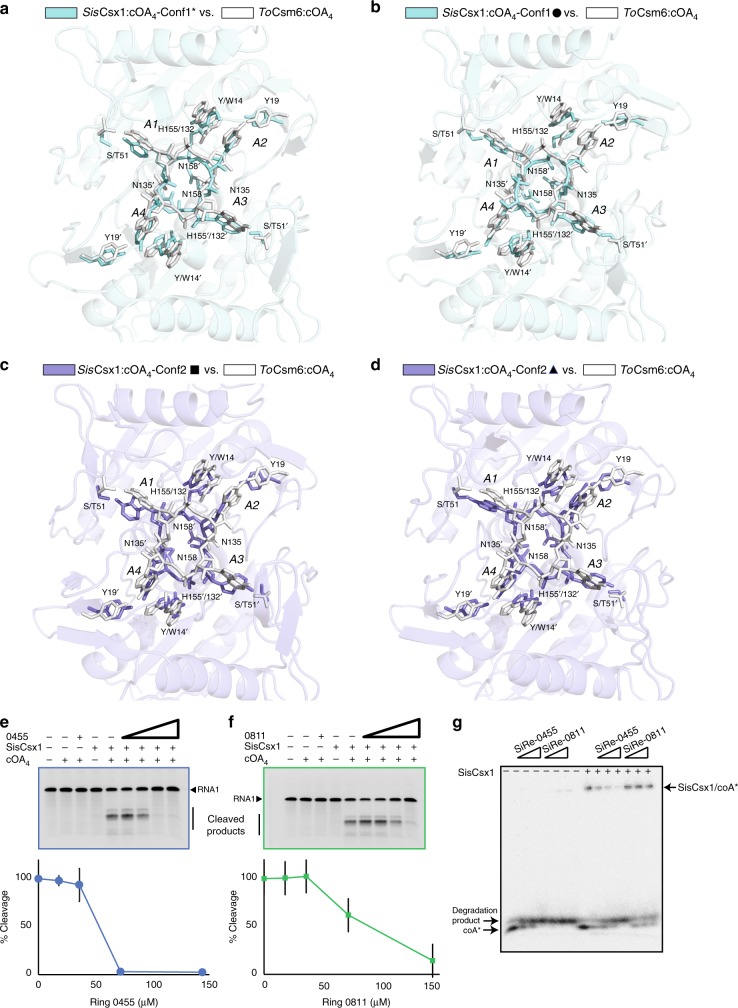
Fig. 4.
Comparison of SisCsx1:cOA4 and ToCsm6:cOA4 CARF domains and RNase deactivation. Detailed view superimposition at the CARF domains of SiScsx1:cOA4 complex in Conf1 asterisk (a) or Conf1 solid circle (b) vs. ToCsm6:cOA4. Detailed view superimposition at the CARF domains of SiScsx1:cOA4 complex in Conf2 solid square (c) or Conformation 2 solid triangle (d) vs. ToCsm6:cOA4. S. islandicus Ring nucleases 0455 (e) and 0811 (f) deactivate the RNase activity of SisCsx1. Increasing concentrations of both Ring nucleases from 10 nM to 150 nM were incubated with 25 nM of cOA4 at 70 °C for 30 min followed by addition of 18 nM of SisCsx1 and 2.5 μΜ of RNA1 and further incubation at 70 °C for 5 min. The reactions were then separated using 15% Novex TBE-urea gel (Invitrogen). g Cleavage of cOA4 by Ring nucleases 0455 and 0811. On the left side of the gel cOA4 is incubated with Ring nucleases 0455 and 0811. The cyclic compound migrates faster than the linear product generated by the ring nucleases. On the right side of the gel SisCsx1 has been included in the reaction showing that it does not cleave cOA4 and protects its degradation by the ring nucleases. Each experiment for the e, f, g panels has been repeated at least three times. The error bars represent the s.d. Source data are provided as a Source Data file for 4E-G