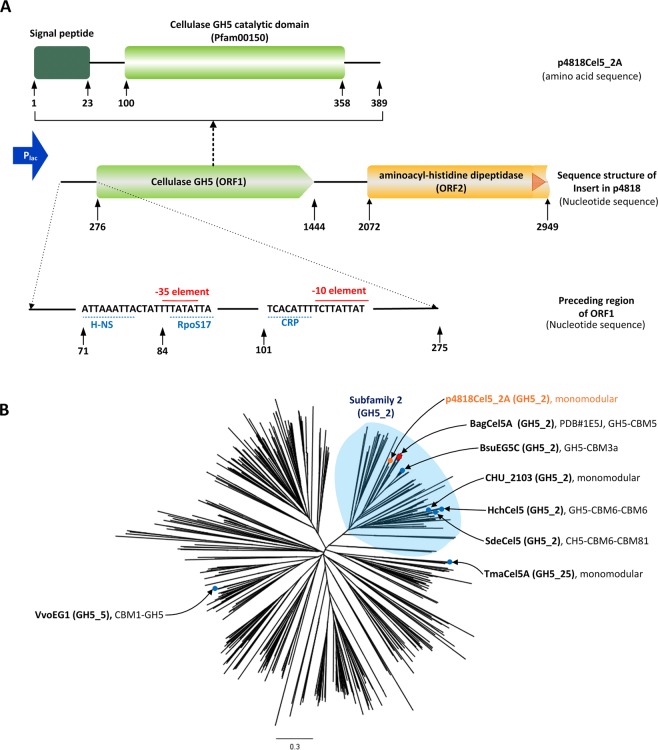
Figure 1.
Organization of the insert from the p4818 positive clone and phylogenetic analysis of p4818Cel5_2A with other characterized members of the GH5 family. (A) Organization of the insert from positive clone of p4818. Two open reading frames (ORFs) were identified within this insert. The predicted −10 and −35 promoter elements are shown in red above the sequences and the identified transcription factor binding sites are underlined in blue. The prediction for bacterial promoters was performed using the online tool of BPROM-Prediction67 against 274 bp sequence proceeding to the cellulase p4818Cel5_2A coding region. Enzyme modules were identified using the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de/). (B) Phylogenetic analysis of p4818Cel5_2A with sequences of characterized GH5 enzymes. All sequences were aligned using MUSCLE in Geneious version 8.0.5. The tree was constructed using Geneious Tree Builder version 8.05. The other reported processive GH5 endoglucanases were indicated by blue dot, and the sequence of template structure (PDB#1E5J) was indicated by red prism.