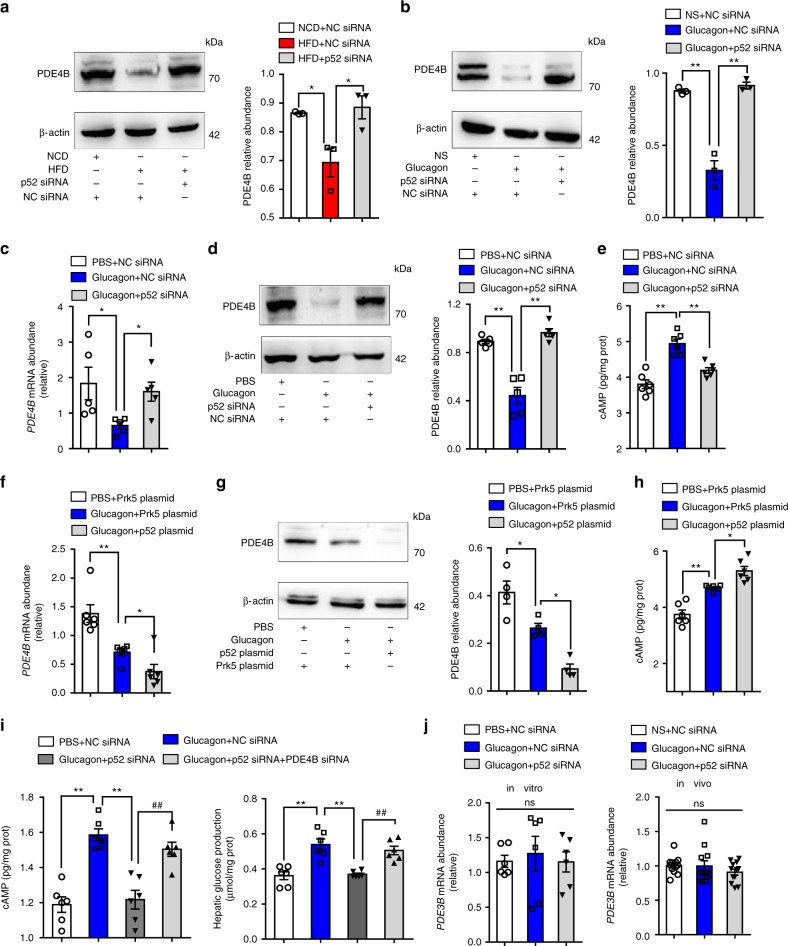
Fig. 3.
p52 inhibits PDE4B expression to promote cAMP accumulation. a, b Liver PDE4B protein expression in HFD-fed (a) and glucagon-stimulated mice (b). Bar graphs represent data normalized to β-actin levels (n = 3). c The mRNA levels of PDE4B in HepG2 cells transfected with p52 or NC siRNA (n = 5). Bar graphs represent the levels of genes normalized to β-actin. d The protein expression of PDE4B in p52 knocked down HepG2 cells, β-actin levels served as loading control (n = 5). e cAMP level in HepG2 cells transfected with p52 siRNA (n = 6). f The mRNA levels of PDE4B in HepG2 cells transfected with p52 overexpression plasmid (n = 5). g Western blotting of PDE4B in p52 overexpression cells (n = 4). h cAMP level in HepG2 cells transfected with p52 overexpression plasmid (n = 6). Bars represent mean ± SEM values. i Intracellular cAMP levels and glucose output in primary hepatocytes transfected with p52 siRNA with or without PDE4B siRNA (n = 6). j Relative mRNA abundance of PDE3B in glucagon stimulated HepG2 cells (100 nM glucagon for 1 h, in vitro) or mice liver tissue (2 mg/kg glucagon for 1 h, in vivo), β-actin levels used as a reference (n = 6). PDE phosphodiesterase, HFD high-fat diet, NS normal saline, ns not statistically significant, PBS phosphate buffer solution. Values represent mean ± SEM. Statistical differences were determined by one-way ANOVA. *p < 0.05 vs. the control group, **p < 0.01 vs. the control group. ##p < 0.01 vs. AAV8-p52 or p52 plasmid group. Source data are provided as a Source Data file