Significance
Many plant pathogenic bacteria can extensively colonize leaf surfaces before entry and multiplication within the leaf to cause disease. While these habitats presumably require distinct adaptations, the genes required in these habitats and how they would differ was unknown. Using a genome-wide library of barcoded insertional mutants in the plant pathogen Pseudomonas syringae, we ascertained the genes required to colonize these habitats. A lack of association between gene expression and a detectable contribution to fitness suggests that many genes that are highly expressed or induced in planta are dispensable or redundant in the environment examined. As a model bacterium for plant pathogenesis and colonization, our comprehensive genetic dataset allows us to better understand the traits needed for association with leaves.
Keywords: epiphytes, endophytes, gene expression
Abstract
The foliar plant pathogen Pseudomonas syringae can establish large epiphytic populations on leaf surfaces before apoplastic colonization. However, the bacterial genes that contribute to these lifestyles have not been completely defined. The fitness contributions of 4,296 genes in P. syringae pv. syringae B728a were determined by genome-wide fitness profiling with a randomly barcoded transposon mutant library that was grown on the leaf surface and in the apoplast of the susceptible plant Phaseolus vulgaris. Genes within the functional categories of amino acid and polysaccharide (including alginate) biosynthesis contributed most to fitness both on the leaf surface (epiphytic) and in the leaf interior (apoplast), while genes involved in type III secretion system and syringomycin synthesis were primarily important in the apoplast. Numerous other genes that had not been previously associated with in planta growth were also required for maximum epiphytic or apoplastic fitness. Fourteen hypothetical proteins and uncategorized glycosyltransferases were also required for maximum competitive fitness in and on leaves. For most genes, no relationship was seen between fitness in planta and either the magnitude of their expression in planta or degree of induction in planta compared to in vitro conditions measured in other studies. A lack of association of gene expression and fitness has important implications for the interpretation of transcriptional information and our broad understanding of plant–microbe interactions.
Many plant pathogenic bacteria are capable of extensive colonization of leaf surfaces before their entry and multiplication within the leaf. As such, epiphytic (leaf surface) populations on asymptomatic plants are considered a reservoir of inoculum that under the appropriate conditions can lead to infection. The likelihood of disease could be predicted from the epiphytic population size of the pathogen several weeks before disease symptoms were observed (1). Therefore, the ability to form large epiphytic populations is a measure of success for such a pathogen, and factors that determine its ability to grow on leaves would be considered fitness factors. After entry into the apoplast, bacterial numbers often increase greatly and disease is associated with those sites in which large internal population sizes have been achieved (2). The ability to grow within the apoplast of plants is thus also a measure of its fitness. In addition to plant pathogens, a diversity of other bacteria and fungi typically colonize the surface of aboveground plant parts. Such commensal bacteria and fungi are, however, typically limited to epiphytic growth with only very small numbers of such taxa found within plant tissue as endophytes (3). It is presumed that the growth of epiphytic bacteria is supported by their consumption of a variety of carbon and nitrogen-containing compounds that transit from the interior of the plant to the leaf surface, or are deposited from external sources, such as aphid honeydew or pollen (4, 5).
A variety of mono- and disaccharides are thought to constitute the majority of the carbon-containing compounds on leaf surfaces, with smaller amounts of other sugars, organic acids, and amino acids also present (6, 7). The absolute amount of such nutrients on leaves is generally low, and the growth of epiphytic bacteria is typically carbon-limited (6). Furthermore, the abundance of such nutrient sources on plants is spatially heterogeneous (4, 6). Because of the apparent chemical complexity, and spatially heterogeneous chemical and physical features of leaves, those traits needed for epiphytic fitness remain largely uncharacterized (8). Only limited descriptions of the chemical and physical environment found within the apoplast of plants have appeared (9). While many of the nutrient resources on the surface are apparently also present in the apoplast, the chemical and physical environment there is largely unknown. Water availability apparently limits intercellular growth (10), and the ability of pathogens to induce plants to release water into the apoplast may be a major feature required for exploitation of this habitat (11, 12). While the apoplast provides bacterial cells protection from environmental stresses on the leaf surface, they are in intimate proximity to living plant cells, and thus subject to inhibitory responses by the plant mediated by the innate immune system (3, 7). Taking these data together, it is clear that a large repertoire of traits beyond resource acquisition, such as motility, habitat modification, and various interactions with the plant may be needed by a plant pathogenic bacterium to exploit both the leaf surface and the leaf interior. Unfortunately, very few such fitness traits beyond those associated with interactions with the plant immune system have been identified.
Pseudomonas syringae is a plant pathogenic bacterial species that includes strains pathogenic on a wide variety of different plant species (13). Most strains have a prominent epiphytic phase, especially on the plant species for which they can also cause infection. Strains of P. syringae are commonly found as epiphytes on a variety of both host and nonhost plants, both in agricultural systems as well as native plant communities (14, 15). Many strains are capable of catalyzing ice formation, and because they can be found in rainfall, pristine snow, as well as in water sources around the world, are thought to play an important role in the water cycle by initiating ice formation central to the precipitation process (15). Such a connection to precipitation may also serve as a vehicle for long-distance dispersal as well as a mechanism for migration to plants after dispersal (16).
The model strain P. syringae pv. syringae B728a (B728a) is a strong epiphytic colonizer that was originally isolated from common bean (Phaseolus vulgaris), and is capable of causing brown spot disease (17). It is the best-studied member within P. syringae phylogroup II. This monophyletic clade contains strains that are overrepresented in environmental samples and are generally better epiphytes than members of other clades (18, 19). In addition, phylogroup II contains many strains with broad host ranges (19). Strain B728a is also pathogenic on Nicotiana benthamiana (20) and pepper (Capsicum annuum) (21). Like other ice nucleation active strains of P. syringae, this strain contributes to frost damage in frost-sensitive plant species by limiting their ability to supercool and avoid damaging ice formation (22). Strain B728a is also a model for phytotoxin production, and contains a much smaller type III effector repertoire than many other strains, such as P. syringae pv. tomato strain DC3000 (23, 24). As such, it has been hypothesized that this lower type III effector repertoire belies an increased reliance on broad-spectrum toxins, as well as ice nucleation ability that contributes to its broad host range and more general environmental distribution (25). This robust epiphytic colonizer and ubiquitous plant pathogen is thus a useful model to examine traits needed for bacterial success in diverse environments.
The genes that are putatively the most ecologically relevant to the success of bacteria on plants have typically been identified on the basis of their transcriptional induction or expression in a given habitat (26). Measurements of gene expression, directly via microarray or RNA sequencing (RNA-seq), or indirectly through reporter genes or in vivo expression technology, have been used in P. syringae to identify genes that have host-responsive expression patterns (27–31). Validation of the role of genes identified by this method, however, usually involves targeted disruption of such genes individually with subsequent assessment of changes in behavior. This is a laborious procedure that cannot be readily applied to the genome as a whole. The high variability of the population size of a given strain after inoculation, especially on leaf surfaces, makes the comparison of population sizes between mutants and parental strains difficult. Large numbers of replicate samples are required to distinguish differential growth of such strains unless they differ greatly (32). Moreover, transcriptional studies may be of limited use in identifying host-colonization genes due to the lack of correlation between gene expression and contribution of those genes to fitness that is often observed in vitro (33). Examples of such a lack of correspondence of gene expression and fitness contribution in some Pseudomonas species on hosts have appeared (34, 35), but it is unclear how prevalent such a lack of connection might be.
To ascertain the roles of individual genes in P. syringae during epiphytic and apoplastic colonization on a genome-wide scale, we utilized a highly parallel transposon-based genomic screen. A variety of techniques taking advantage of high-throughput sequencing have been used to identify genes contributing to host colonization (36, 37). For example, genomic comparisons can reveal differential gene abundance in strains and genes that are under putative positive selection, suggesting that they might be contributing to host-specific fitness (38–40). However, confirmation of the role of such genes typically requires laborious mutation analysis, as discussed above. Random mutagenesis techniques can enable genome-wide, gene-specific fitness contributions to be measured in a given habitat. One such strategy employs transposon sequencing (TnSeq) wherein the relative proportion of a transposon mutant in a given gene within a mixture of such mutants is assessed both before and after the strain mixture experiences a given condition. The number and genome location of the mutants in the mixture is determined by determining the sequences adjacent to the transposon by high-throughput sequencing in each experiment (41). Recently developed random-barcoded transposon sequencing (RB-TnSeq) (42) is a modification of TnSeq that enables a transposon library to be used more easily for multiple assays since tagged transposons are used in mutagenesis and need be mapped only once. Eliminating the need to remap transposon insertions in each experiment dramatically reduces the effort to carry out fitness screens in multiple conditions by using a single library of mutants. In a transposon mutant pool where each transposon is linked to a unique 20-nucleotide barcode, insertion mutant fitness is calculated through amplicon sequencing of the barcode regions to calculate the relative abundance of a given strain. Change in barcode relative abundance over time is used as a proxy for strain fitness within the population. RB-TnSeq was recently used to identify genes required by Pseudomonas simiae for its invasion of Arabidopsis thaliana roots (43). In this study, we used RB-TnSeq to identify genes in P. syringae needed for its colonization of both the surface and interior habitats of bean. Since the stimulon for these 2 habitats had previously been determined (15), we also addressed the extent to which transcriptional changes in gene expression were predictive of the fitness contributions of these same genes.
Results
Adapting RB-TnSeq for an Epiphyte and Foliar Pathogen.
In order to screen for genes in P. syringae strain B728a contributing to host colonization, we generated a randomly DNA-barcoded mariner transposon library using the Escherichia coli donor library created by Wetmore et al. (42). The sequenced B728a mutant library consisted of 281,417 strains with insertions that map to the B728a genome, each containing a unique DNA barcode. Computationally removing insertional mutants outside the central 10 to 90% of coding region of a given gene resulted in 169,826 genic strains for analysis, with a median of 21 insertions per gene. The number of usable insertional mutants for each gene was correlated with the number of TA dinucleotide sites within each coding region (Pearson correlation coefficient r = 0.71) (SI Appendix, Fig. S1). We analyzed fitness contributions for 4,296 of 5,137 (84%) protein-coding genes that harbored sufficient insertions for analysis.
The rich medium King’s B (KB) was used for library recovery prior to plant inoculations, so overnight growth in this condition was used as the control against which growth of the mutants on the leaf surface and in the apoplast (SI Appendix, Fig. S2) was compared. All experiments analyzed herein passed quality-control metrics that were previously established for in vitro studies in Wetmore et al. (42). The requirements for a successful experiment include ≥50 median reads per gene and consistency in the calculated fitness estimate obtained from mutants with insertions in the 3′ and 5′ half of a gene (42).
For each experiment, an aliquot of the mutant library was grown to midlog phase in KB (approximately 5 generations) and a sample of the library was taken immediately before inoculating either the surface or interior of plants (time0). After growth in each condition, cells were recovered from either the surface or interior of the plants and prepared for sequencing. Fitness for each strain was calculated as the log2 of the ratio of barcode abundance following growth in or on plants with that barcode abundance obtained initially at time0. Gene fitness is calculated as the weighted average of the individual strain fitness values (42). Insertions in the majority of genes did not change fitness as measured by relative barcode abundance in the population, and thus the fitness values for most genes were close to 0.
Identification of the Essential Gene Set of B728a.
For 920 genes, fitness could not be calculated due to a lack of sufficient insertional mutants. Of those, only 5 do not contain TA dinucleotide sites and thus are not accessible by mutagenesis with the mariner transposon we used. Of these genes, 512 did contain at least 1 mapped insertion, but we were unable to calculate fitness values for them due to the small number of sequencing reads at time0 (SI Appendix, Fig. S3), suggesting that they were relatively unfit in vitro compared to other mutants, and thus in low relative population size in the library. Based on analysis of the TnSeq data, we predicted at least 392 genes to be essential or nearly essential for B728a growth on Luria-Bertani (LB) agar, as they contain numerous TA sites but do not contain any mapped insertion strains in our library. The 392 predicted essential genes include many annotated as being involved in translation (including tRNAs), energy generation, and cofactor metabolism (SI Appendix, Table S1). Using the Integrated Microbial Genomes (IMG) database (44), we identified orthologs in Psudomonas aeruginosa PAO1, P simiae WCS417, and Pseuodomonas stutzeri RCH2 for 2,349, 2,560, and 1,450 B728a genes, respectively. Based on predicted essential genes in these strains (45–47), we identified the degree of overlap in predicted essential or nearly essential genes for growth on LB (SI Appendix, Table S2); 181 genes are predicted to be essential or nearly essential in all 4 strains examined here. Dataset S1 contains the list of all B728a genes with their predicted orthologs and whether those orthologs were originally predicted to be essential or nearly essential. Of those genes with predicted orthologs, 48 to 133 were annotated as essential in B728a and nonessential in the comparison strain, or vice versa.
Disruption Mutants with Fitness Defects in Rich Medium.
The pooled library was initially generated in LB but inoculum for the experiments was grown on KB. While these are both rich media, the presence of yeast extract in LB apparently provides resources not found in KB, and thus the growth of certain biosynthetic mutants was limited in KB. We identified 20 genes that were required for maximal growth in KB medium, 9 of which are involved in cofactor metabolism (SI Appendix, Table S3). Of these 20 genes, 13 contributed to fitness in or on plants. For example, the ability to synthesize the cofactor biotin was required for growth in KB, suggesting its limitation compared to other components, such as amino acids and carbon compounds in this medium. However, the presence of biotin auxotrophs in the library indicates that these mutants were able to grow in LB. This suggests that cofactors such as biotin are lacking in KB medium, as well as in the in planta habitats.
Maintaining Genetic Diversity of the Transposon Library In Planta.
A major challenge for the use of complex mixtures to study the relative fitness of component strains in any experiment, especially studies done in planta, is to ensure that all of the mutants in the mixture are well represented after inoculation so as to avoid bottleneck effects. The strength of saturated transposon mutagenesis methods lies in internal replication: The contribution of each gene is assessed by interrogation of the behavior of multiple independent insertional mutant strains. A loss of diversity at the time of inoculation reduces the statistical power for analysis of a given gene. We aimed to maximize the total number of inoculated bacterial cells to maintain population diversity, while achieving a sufficiently low initial inoculum in or on plants so that substantial, competitive growth of the mixture could be assured. Given that the mutant library contains over 200,000 distinctive members, we inoculated a total of ∼107 cells either into or on a collection of plants. So as to achieve a sufficiently low initial population size (approximately 105 cells per gram) that would enable a maximal number of generations to occur in and on leaves over which growth defects could be estimated, we chose to inoculate a large number of leaves (100 pots of plants). Although we observed slight bottlenecks given the concentration of inoculated cells we used, particularly in apoplastic conditions, these samples provided sufficient reads for most mutants to enable analysis of the fitness contribution of nearly all genes (SI Appendix, Table S4); more than 80% and 68% of the unique barcoded mutants were retained in studies of epiphytic and apoplastic growth, respectively. More than 99% of the unique barcoded mutants in the library were retained during in vitro experiments.
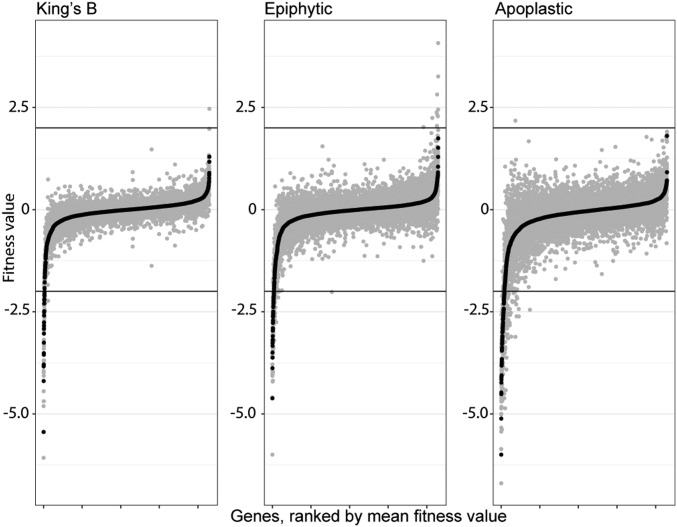
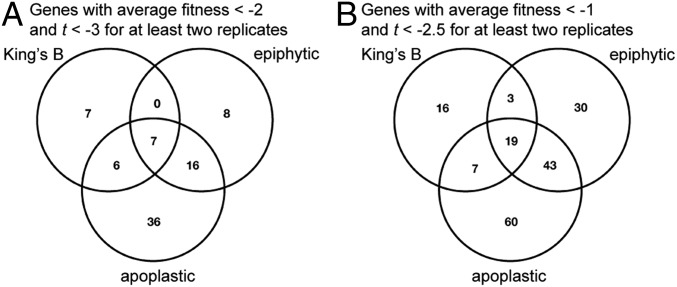
During the growth of strain B728a on leaf surfaces for 2 d, the total number of cells increased ∼100-fold to 107 CFU/g (27), corresponding to 6 to 7 population doublings. Similarly, during growth in the apoplast for 6 d, population size increased about 1,000-fold to over 108 CFU/g (SI Appendix, Fig. S4), indicating at least 10 cell divisions. In theory, in an experiment on leaves in which most strains exhibited 6 generations of growth, mutants completely incapable of growth should exhibit a fitness of about −6 (42). In practice, insertions in very few genes exhibited such an extreme lack of fitness (Fig. 1). While several genes, such as those conferring auxotrophy upon disruption, had large negative fitness values in planta, these values were less than that of the number of generations to which the population as a whole underwent, perhaps due to the consumption of rare nutrients or to residual cellular resources from the period of population recovery. Eighty genes were identified as contributing strongly to growth, in which the corresponding mutants exhibited fitness values less than −2 (exhibiting only 25% as much growth in the population relative to that of the typical strain in the mutant population) (Fig. 2A). Mutants in an additional 69 genes exhibited fitness values less than −1 but greater than −2 (Fig. 2B), suggesting that these genes contributed somewhat less to fitness (42). We did not normalize fitness values by the average number of generations in a given experiment, as these values are difficult to estimate and likely vary by plant within an experiment.
Fig. 1.
Rank-ordered mean gene fitness values for each condition in which P. syringae was grown. Fitness values for independent replicate experiments are shown in gray, while mean fitness values are plotted in black. Gene-fitness values are calculated as the log2 ratio of the barcode counts following growth in a given condition compared to the barcode counts from midlog phase cultures following library outgrowth and before inoculation. Fitness values are normalized across the genome so the typical gene has a fitness value of 0. Black lines at fitness values of −2 and 2 are used to indicate strong phenotypes; for example, a value of −2 indicates that mutants in that gene were 25% as fit as the typical strain in the mutant library. In each dataset, fitness values less than −2 or greater than 2 are more than 3 SDs from the mean (∼0).
Fig. 2.
Genes with significant contributions to competitive fitness in the experimental conditions tested. (A) Venn diagram of genes with average fitness values less than −2, and t < −3 for at least 2 experimental replicates. (B) Venn diagram of genes with average fitness values less than −1, and t < −2.5 for at least 2 experimental replicates.
Despite the differences in the number of generations in epiphytic versus apoplastic growth in plants (27), we observed similar ranges in overall fitness values for individual genes in these 2 habitats (Fig. 1). Mean fitness values ranged from −4.6 to +1.7 (epiphytic) and −6.0 to +1.8 (apoplastic). For each gene, we averaged fitness values for the 2 replicate growth experiments in KB and the 3 epiphytic and 3 apoplastic experiments performed. We focused our analysis on genes contributing most strongly to fitness: Those for which mutants had an average fitness less than −2 and for which the t-score was less than −3 in at least 2 replicate experiments. Since the plant host constitutes a more variable environment than most in vitro experiments, and expecting that many genes would not individually make large contributions to fitness, we also examined genes with fitness values less than −1 but with t <−3. Special attention was placed on those genes with such values that are operative in a given metabolic pathway or could be placed in the same functional category. Approximately 50% of all genes exhibiting fitness values less than −2 or −1 in either epiphytic or apoplastic habitats were verified in at least 2 of 3 replicate experiments (SI Appendix, Fig. S5).
Genes Required Specifically for Colonization of the Leaf Surface.
We identified 31 genes that were highly important for fitness on the leaf surface (SI Appendix, Table S5), although all but 8 were also important in the apoplast. Among these 8, genes in the predicted operon Psyr_2461-2 had a particularly strong epiphytic phenotype, with average fitness values of −2.1 and −3.2. Psyr_2461 is a hypothetical protein containing a domain of unknown function (DUF934) and Psyr_2462 is homologous to the sulfite reductase cysI in P. aeruginosa. Glutamate synthase (NADPH) subunit genes gltB (Psyr_0411) and gltD (Psyr_0412) also contributed strongly to epiphytic growth, having average fitness values of −2.0 and −1.1. Disruption of the putative phage-related protein Psyr_4512 also strongly reduced epiphytic fitness (average fitness value = −2.1).
Genes Contributing Specifically to Colonization of the Leaf Apoplast.
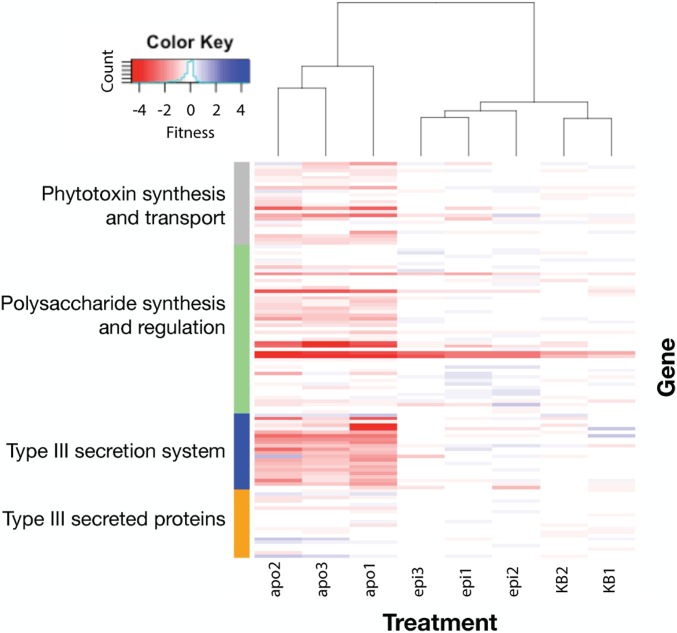
Disruption of many genes encoding known virulence factors, including those in the type III secretion system (T3SS) (SI Appendix, Fig. S6) and phytotoxin biosynthesis genes greatly reduced the growth of P. syringae in the apoplast. Of the 65 genes that were highly important (average fitness less than −2) for apoplastic colonization (SI Appendix, Table S6), 36 were important in this habitat but not on leaf surfaces. The T3SS genes we observed as essential for successful apoplastic colonization are exclusively involved in the T3SS machinery, as transposon insertions in most individual effector genes generally had no fitness phenotype (Fig. 3). Of the secreted type III effectors, hopAB1 had the largest negative average fitness value (−0.55). While the fitness contribution of this gene was less than many others, growth of mutants in this gene was decreased in all 3 experimental replicates (SD = 0.065, t < −3.5 for all). As t-scores are positively correlated with measures of fitness, the low variance in fitness seen among the 21 insertional mutants for this gene provide confidence in the rather modest fitness estimates for this gene.
Fig. 3.
Fitness contributions of genes involved in phytotoxin synthesis and transport, the T3SS, and polysaccharide synthesis and regulation are required for apoplastic colonization. Gene fitness values are shown from 3 replicate epiphytic experiments (epi), 3 replicate apoplastic experiments (apo), and 2 replicate experiments in KB medium. An expanded version of this figure containing gene names and loci can be found in the SI Appendix. The treatment ordering and corresponding dendrogram are calculated based on these genes only.
Production and secretion of the phytotoxin syringomycin was required for competitive fitness in the apoplast. The syringomycin regulator syrP and syringomycin efflux transporter syrD both had large apoplastic-specific phenotypes when disrupted. In contrast, syringopeptin and syringolin mutants did not have significant apoplastic fitness defects in our experiments. Polysaccharide synthesis and regulation was highly important for competitive fitness in the apoplast. Mutants in alginate regulation (algU) and biosynthesis were dramatically less competitive than the typical mutant in the library. Group 1 glycosyltransferase encoding genes (Psyr_0920 and wbpYZ) also contributed substantially to apoplastic growth (Fig. 3).
The 2-component system GacA/GacS was moderately important in the apoplast (average fitness = −0.9 and −1.5), but interestingly, their disruption resulted in increased fitness on the leaf surface (average fitness = 1.3 and 1.7). Conversely, glutathione synthase (gshB) was important in KB (average fitness = −1.5) and on the leaf surface (−1.2), but disruption of this gene increased competitive fitness in the apoplast (+1.8). Generally, however, insertional mutations rarely significantly increased fitness in any experiment.
Genes Required for the Colonization of Both the Leaf Surface and Apoplast.
Overall, the categories of “amino acid metabolism and transport,” “polysaccharide synthesis and regulation,” and “nucleotide metabolism and transport” were enriched in genes with average fitness less than −2 in both epiphytic and apoplastic habitats relative to that in rich medium (SI Appendix, Table S7). We identified 31 genes that were highly important for epiphytic colonization, and 65 genes that contributed to apoplastic growth. Approximately one-third of all genes contributing to leaf colonization were also important in the apoplast (Fig. 2).
Genes required for the biosynthesis of several different amino acids were highly important in colonization of both the leaf surface and the apoplastic space. Genes required for biosynthesis of tryptophan, proline, and the shared biosynthetic pathway of isoleucine/leucine/valine were among those with the largest contributions to fitness in both in planta conditions tested. Additionally, genes involved in biosynthesis of methionine were important for epiphytic survival, as seen previously (48), and disruption of these genes caused even greater decreases in apoplastic growth, suggesting that these resources are in low abundance in these habitats. For example, average fitness values for metW and metZ auxotrophs were less than −4 in the apoplast, but approximately −0.8 on leaf surfaces. A similar, albeit less dramatic, pattern of proportionally larger requirements for histidine biosynthesis under apoplastic growth was also seen. The production of cofactors, such as pantothenate (vitamin B5) requiring panC, was important in both in planta conditions but contributed more to the growth on the leaf surface. Genes involved in nucleotide biosynthesis (purFL) were also highly important for growth both in and on leaves.
The genes mdoGH encoding glucan synthesis were required for optimal growth on both the leaf surface and in the apoplast. Hypothetical proteins encoded by Psyr_0532, Psyr_2461, and Psyr_4158 (eftA) all made significant contributions to fitness both epiphytically and in the apoplast. Psyr_0532 contains a group 1 glycosyltransferase domain.
Validation of Fitness Estimated in Disruption Mutant Mixtures with Targeted Deletion Strains.
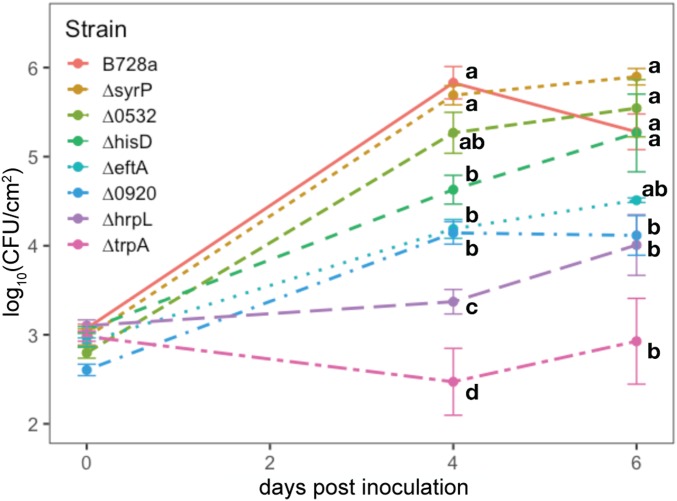
To determine whether the growth deficiencies of mutant strains in the pooled assays were predictive of that when grown in isolation, we constructed targeted deletion mutants of several genes that contributed differentially to apoplastic fitness of P. syringae (SI Appendix, Figs. S7–S9). Amino acid auxotrophs ∆trpA (average apoplastic fitness = −2.7) and ∆hisD (average fitness = −3.0) were inoculated individually into the apoplast. While ∆trpA was almost incapable of growth, the population size of ∆hisD was about 10-fold lower than the WT strain 4 d after inoculation. Similarly, while a ∆hrpL type III secretion mutant (average fitness = −2.2) achieved an apoplastic population size that was only about 1% that of the WT strain, the population size of a ∆syrP mutant deficient in production of syringomycin (average fitness = −2.1) was only slightly lower than that of the WT strain when inoculated separately into plants (Fig. 4).
Fig. 4.
Apoplastic growth of B728a and deletion strains in bean. Growth of the amino acid auxotrophs ∆trpA and ∆hisD, type III regulatory mutant ∆hrpL, and syringomycin mutant ∆syrP. Apoplastic fitness of deletion mutants of glycosyltransferase genes Psyr_0532 and Psyr_0920, and hypothetical protein eftA. Error bars represent the SEM. At each time point, strains labeled with the same letter are not significantly different (Tukey’s honest significant difference test, P < 0.05).
We also assessed fitness of directed mutants of 3 poorly understood genes, 2 of which are predicted group 1 glycosyltransferases, that contributed substantially to competitive apoplastic growth. Deletion mutants of ∆eftA (average fitness = −1.4, a hypothetical protein), and ∆Psyr_0920 (average fitness = −2.4, a group 1 glycosyltransferase) both achieved apoplastic population sizes that were 10-fold lower than that of the WT strain when assessed both 4 and 6 d after inoculation. In contrast, the apoplastic population size of ∆Psyr_0532 (average fitness = −1.6, a hypothetical protein containing a group 1 glycosyltransferase domain), was only slightly less than that of the WT strain (Fig. 4).
Fitness Contributions of Genes Do Not Correlate Well with Their Level of Transcriptional Expression or Inducibility in or on Plants.
We compared previously published global transcriptional patterns for the genes in strain B728a when grown on leaf surfaces and in the apoplast (27), as well as for genes in the related strain DC3000 grown in the apoplast of Arabidopsis (28) with that of the fitness contributions of the genes measured here to determine how predictive gene expression was to the functional role of these genes in growth in and on leaves. Both the absolute levels of gene expression in various in planta conditions, as well as that of the changes in expression of a given gene in planta relative to that in cells grown in a minimal medium (MM, for B728a) or KB (for DC3000), were used as predictors. In general, while many genes exhibited substantial elevated or depressed expression on or in plants compared to that in culture medium, disruption of these same genes often had little or no impact on the competitive fitness of the mutant strain in this study (Fig. 5). For example, while many amino acid auxotrophs were significantly less fit on the leaf surface and leaf interior, expression of biosynthesis genes for tryptophan, histidine, proline, and methionine was not induced, and instead was repressed, in these habitats compared to in vitro conditions. Similarly, while genes involved in biosynthesis of the cofactor pantothenate (shared with biosynthesis of the branched amino acids) were required for competitive fitness, their expression was down-regulated in planta. Likewise, although the expression of genes encoding several hypothetical proteins were strongly increased in planta, suggesting that they may play an important role in growth on plants, their disruption had little effect on the competitive fitness of these mutant strains. Exceptions to this lack of association between gene induction and contribution to fitness are the genes (syrP and syrD) required for the biosynthesis of syringomycin in the apoplast; these genes were highly up-regulated in planta and mutants in this gene cluster also were much less fit. Similarly, scrB, involved in sucrose metabolism, is strongly up-regulated specifically in the apoplast and was also specifically required for competitive fitness in that environment. Generally, however, examples of genes in which a concordance between absolute or plant-dependent levels of transcription and their fitness contribution in planta were rare.
Fig. 5.
The magnitude of fitness contributions of genes in P. syringae do not correlate well with their absolute level of expression (Upper) or fold-change of these genes in planta compared to that in hrp-inducing MM or KB (Lower). Epiphytic (Left) and apoplastic (Center) gene expression in B728a is a measure of fluorescence in microarrays (27). A comparison of B728a apoplastic gene fitness to expression of orthologs in the closely related strain DC3000, measured in A. thaliana, reveals a similar trend (Right). DC3000 gene expression is calculated as the number of reads per million (28). Fold-change in gene expression was calculated as a log2 of the ratio of gene expression estimated from microarray fluorescence (B728a) or RNA-seq (DC3000) in planta relative to that in MM or in KB (27). The Pearson correlation coefficients are shown.
Discussion
Competitive colonization assays are a very sensitive method by which differences in relative fitness can be assessed. In a phenotypically heterogeneous population, changes in relative proportion of a given member provide a direct assessment of relative fitness. A notable exception that would preclude such a process would be one in which there is the production of shared goods (such as siderophores) that can be coopted by nonproducers, as predicted by the Black Queen Hypothesis (49). Random mutagenesis methods, such as TnSeq, in which insertional mutant strains are grown in large pooled mixtures, have the advantage of identifying conditionally important genes in a genome-wide manner. By being intrinsically parallel in their structure, the ability to readily distinguish and enumerate each of the individuals in such a mixture by RB-TnSeq provides both a high-throughput and highly sensitive means by which relative fitness of the individual strains can be assessed. Furthermore, the creation of multiple independent insertional mutants for each gene provides substantial internal replication, increasing confidence in the fitness phenotype quantified for any given gene. An advantage that RB-TnSeq has over more classic TnSeq is that the association of a given transposon insertion within a gene need be done only once, since a random DNA barcode can then be unambiguously associated with that insertional event thereafter. Such a process then allows the use of the same barcoded transposon library for multiple experiments by simply sequencing and enumerating the DNA barcodes, enabling repeated interrogation of the role of the genes in a species such as P. syringae in many different environmental settings. The utility of RB-TnSeq has been demonstrated by its application to a myriad of different bacterial species exposed to hundreds of distinct environmental settings, enabling functions to be assigned to many previously uncharacterized genes (47). Our demonstration of the utility of RB-TnSeq in this study should enable us and others to greatly expand the association of genes in P. syringae to the myriad of functions in which it might be expected to participate, in the many chemically and physically different habitats that it colonizes.
P. syringae is a robust colonizer of both leaf surfaces and the apoplastic space of the host plant common bean. In these habitats, this strain exhibited sufficient growth to enable RB-TnSeq to quantify the contribution of individual genes that directly contribute to competitive fitness in a heterogeneous population. It would be expected that the ability of such a method to resolve differences in fitness contributions of these genes would increase with the number of generations of growth that the population of mutants would have undergone during an experiment. Given the large number of genes for which some fitness contribution could be measured, we focused our analysis here on those genes having the largest fitness contribution. Furthermore, there is generally higher statistical support for the validity of fitness estimates for those genes contributing more to fitness (SI Appendix, Fig. S6), given that they were large and reproducible across replicate experiments. Genes associated with somewhat lower, but consistent, fitness values (SI Appendix, Table S8) are likely also biologically significant, and future studies can explore the roles of these genes during in planta growth in more depth. In the present study the high internal replication intrinsic to the barcoded transposon library, and the use of several replicate experiments for each in planta condition, has provided a compelling list of broadly important genes for further analysis.
Transposon-based approaches have been useful in identifying essential bacterial genes in other taxa, although our knowledge of essential genes in Pseudomonas species is limited. Analysis of a 100,000 strain insertional library in P. aeruginosa identified 336 of the 5,606 genes to be essential in LB (46). Recent work has identified 473 genes as likely to be essential in P. simiae, 430 in P. stutzeri, and 325 to 442 in Pseudomonas fluorescens (depending on the strain) (47). We identified 392 genes to be likely essential for B728a growth in LB, comprised of functional categories generally seen to be essential in diverse bacteria. Given that we could calculate fitness contributions for 84% of the protein-coding genes in the environmental conditions tested here, the proportion of genes found to be essential in P. syringae appears similar to that in E. coli and P. stutzeri (42). Differences of essential gene predictions between related strains highlight the value of making essential gene predictions using multiple conditions or strains, such as this TnSeq study of P. aeruginosa grown on 6 different media (50).
The identification of genes involved in anabolic processes, such as cofactor production and amino acid biosynthesis that contribute to the fitness of P. syringae in a given habitat, provides some insight into the availability of such resources in that setting. This logic of anabolic mutants as reporters of habitat resources provides insight into the resources on the surface and in the intercellular spaces of plants. The finding of fitness defects for many amino acid biosynthetic genes is a clear example of this concept. The much lower fitness of auxotrophs for several different amino acids suggests an acute limitation of these essential metabolites both on the leaf surface and in the apoplast. Genes within the biosynthetic pathway of tryptophan had the largest effect on fitness when disrupted in our study, both on the leaf surface and in the apoplast. This is consistent with a previous study showing tryptophan synthesis as critical for both epiphytic and apoplast growth of B728a (51). Similarly, amino acids such as methionine and histidine are present in low levels in the apoplast, imposing a need for their synthesis by bacterial colonists (9, 52). Similar to these observations, biosynthetic genes for tryptophan (and its precursor anthranilate) were identified as important for fitness in a TnSeq study examining Pantoea stewartii colonization of maize xylem (53). A TnSeq screen in Dickeya dadantii in rotting plant tissue also noted a significant decrease in competitive fitness in planta for leucine, cysteine, and lysine auxotrophs that could be negated through the external addition of amino acids (54). Since most amino acids are apparently present at relatively low concentrations in the bean apoplast (9), it could be expected that many auxotrophs are incapable of growth without the ability to synthesize these essential nonsubstitutable metabolites, a model supported by the observations of this study.
Unlike certain anabolic genes, those involved in catabolism typically had more subtle phenotypes when disrupted. This is likely due to the presence of diverse and substitutable carbon and nitrogen sources, such as sugars and organic acids in and on plants (9). It was noteworthy that the fitness of mutants in which sucrose 6-phophate hydrolase encoded by scrB was disrupted was lower in the apoplast (fitness value −1.7). Such an observation is consistent with sucrose being the most abundant sugar in intercellular spaces (9). On the other hand, the genes involved in the metabolism of compounds of lesser abundance that that are not essential would be expected to contribute somewhat incrementally to the fitness of P. syringae in or on leaves. While carbon availability appears to limit the growth of bacteria such as P. syringae on leaves (6) and might also limit the growth of this species in the apoplast, it might be expected that these various compounds represent substitutable resources (9), and that any given compound would contribute relatively little to the overall growth of such a strain if many were present in similar concentrations. In support of this conjecture, while small fitness defects were observed for several mutants harboring disruption of genes essential for catabolism of nutritive compounds, the magnitude of these fitness defects was usually low.
Many genes involved in polysaccharide synthesis and regulation were highly important in leaf colonization. The lipopolysaccharide found in the outer membrane of gram-negative bacteria is known to induce the innate immune response of plant and animal hosts, yet it is required for bacterial stress tolerance in diverse environments (55). Many of the hypothetical proteins having strong plant phenotypes when disrupted here contain glycosyltransferase domains. We hypothesize that these hypothetical proteins contribute to the biosynthesis of O-antigen that decorates LPS, and thus might be involved in camouflaging the cells so as to not be perceived by plant surveillance systems. O-antigen is an essential virulence factor for P. aeruginosa in its colonization of animal tissues (56). While O-antigen is dispensable for growth in culture, it has been recently shown to delay the host immune response during Xylella fastidiosa colonization of grape xylem (57). Alternatively, glycosyltransferase activity may contribute to flagellar modifications in order to avoid plant recognition (58–60). However, this is unlikely to be the major role of these selected genes in P. syringae since known flagellar glycosyltransferases in P. syringae (fgt1 and fgt2) located adjacent to flagellar biosynthesis genes did not measurably contribute to competitive fitness in planta in this study. Nonetheless, we show that 8 genes containing glycosyltransferase domains made large individual contributions to host colonization, suggesting that there may be other important targets for such modification.
Biosynthesis of the exopolysaccharide alginate contributed strongly to growth in the apoplast but not on leaf surfaces. While alginate had been shown to contribute to epiphytic fitness and thus to subsequent disease severity of P. syringae (61, 62), the apoplastic colonization of mutants was not distinguished from epiphytic growth in those studies. The apoplast is thought to be a water-limiting environment for endophytic pathogens (10, 12, 27). Furthermore, the transcriptional activation of the key alginate biosynthetic enzyme algD is induced by high osmolarity (63), and thus alginate biosynthesis would be expected to contribute to fitness in the apoplast, as observed here. While the AlgU regulon includes genes involved in additional stress-responsive processes—such as osmoprotectant transporters (OpuC and Cbc), antioxidant enzymes (KatE, SodC, and CpoF), and the type 6 secretion system (64)—we did not observe any fitness decrease for insertional mutants in those genes. In Pseudomonas putida, alginate production is required for biofilm-mediated survival under desiccating conditions (65). We did not see a significant role of alginate on the leaf surface, potentially due either its potential to be a shared good or to the high humidity used for these epiphytic studies that negated any benefit of water absorption. However, alginate biosynthesis was a clear virulence factor in the apoplast.
Our screen highlighted the fitness role of many known virulence factors, including the T3SS and the phytotoxin syringomycin. It was encouraging to detect T3SS mutants, indicating the bacterial apoplastic densities were not high enough to complement these mutants in trans, perhaps because of the low spatial density of the inoculated cells within the plant. Individual secreted effector proteins did not generally contribute measurably to apoplastic colonization, while mutations in type III pilus genes significantly decreased fitness. This supports existing dogma, whereby type III effector proteins are individually dispensable and collectively essential (66). HopAB1, a secreted type III effector that we found to have the largest contribution to apoplastic fitness among all secreted effectors, has been shown to have a measurable contribution to B728a growth in the bean apoplast (20). While phytotoxin production in strain B728a has been shown previously to induce symptom formation, there has not been compelling data showing a contribution to bacterial growth in plants (67). It is interesting that the genes involved in syringomycin and syringopeptin have strong negative fitness values in our study, suggesting that mutants in these pathways are impaired in growth relative to WT. Our results support the model that P. syringae strains such as B728a with relatively fewer type III effectors have an increased reliance on phytotoxin production for growth in the apoplast (23, 68). The biosynthetic gene cluster for syringomycin also was distinctive in that it was among the few genes that are both up-regulated in planta (15) and contribute to fitness. On the other hand, genes for syringolin biosynthesis, while up-regulated in the apoplast (27), did not contribute to apoplastic fitness in our study. Syringolin contributes to virulence through host proteasome inhibition, which has been shown to counteract stomatal innate immunity (69). Therefore, the role of syringolin is likely limited to the transition from epiphytic to apoplastic growth, a process that was not tested here. Syringopeptin, which is also up-regulated in the apoplast (27), contributed to apoplastic fitness to a much lesser extent than syringomycin. Syringomycin and syringopeptin have the same mechanisms of action, creating membrane pores and causing ion leakage (70). It is unclear why B728a produces 2 seemingly redundant phytotoxins, although it has been proposed that their differential antimicrobial activities contribute to epiphytic survival (71). Both syringomycin and syringopeptin contribute to virulence on cherry (70) and lysis of tobacco protoplasts (71). Since we observed a much larger contribution to bacterial fitness in the bean apoplast from syringomycin than syringopeptin, it is tempting to speculate that the functions of these phytotoxins in virulence may be somewhat host-specific.
Despite the dogma that gene expression is fine-tuned to the metabolic demands of a cell, recent studies of gene expression have shown such regulation to be suboptimal for many bacterial species (33). Despite classic examples of biosynthetic pathways in E. coli having adaptive regulation, many genes in diverse bacteria show little correlation between when they are important for fitness and when they are most highly expressed (33). For example, constitutive expression and regulation by growth rate are common indirect gene regulation strategies that occur for genes with diverse functions and yet are often suboptimal in the laboratory and presumably also in natural environments (33). In P. aeruginosa wound infections, gene expression was also not well correlated with gene contributions to fitness (35). A proposed explanation for such incongruence was that given that P. aeruginosa is considered an opportunistic pathogen that might not have evolved primarily in association with mammalian tissues, its patterns of gene expression might have optimized fitness in very different settings (35). Moreover, in persistent, long-lasting infections, such as the cystic fibrosis lung, adaptive changes in global patterns of gene expression in P. aeruginosa have been observed over time (72), reflecting adaptation to this new habitat.
While P. syringae is a model plant pathogen, it is also commonly observed in many other environmental settings (19). The conditions that the cell would experience on the surface of the plant are likely to be quite different from those in the apoplast (3, 13). Here, we see no correlation between gene expression (either absolute or relative change) and contribution to fitness in the host. It was surprising that genes that were highly expressed or highly induced in cells in or on leaves did not make large contributions to the fitness of the strain. Studies of gene expression are very common, and tend to focus on those genes that are induced in a given environment or condition. However, calculations of inducibility are inherently relative to the reference condition, often a rich or minimal medium. As the calculations of B728a gene induction in planta was performed relative to hrp-inducing MM, we reasoned that some genes would perhaps already be induced. To get around this, we also compared our fitness data to gene-expression measurements in the closely related strain DC3000 growth in planta relative to the more typically used rich culture medium. While the timing of sampling of RNA from the apoplast for these comparative studies was somewhat earlier in the infection process [Yu et al. (27) sampled bacterial cells 2 d postinoculation and Nobori et al. (28) sampled after 6 h, while we sampled after 6 d], we would not have expected temporal changes in gene expression to overwhelm any context-dependent patterns of gene expression. Despite the different conditions used as reference points, the overall trends were similar between these 2 strains in planta relative to the contributions of those genes to fitness measured here. Similarly, many genes that were either weakly expressed or uninduced on or in plants proved particularly important for fitness in these habitats. This lack of congruence can be explained by the fact that many genes are involved in catabolic processes wherein individual pathways would be expected to contribute only incrementally to the success of a strain. Genes for anabolic pathways, on the other hand, might prove essential irrespective of how highly expressed they are. There remain many genes for which the lack of linkage between expression and contribution to fitness remain unexplained. It is evident that directly measuring the contribution of a gene to fitness in different environments is a necessary complement to global transcriptional profiling to understanding the function and behavior of a cell in a given setting.
Although P. syringae is a ubiquitous species, it is most commonly studied in its agriculturally relevant, disease-susceptible plant hosts. Random mutagenesis studies typically observe that a majority of genes in the genome are dispensable, as seen in the relatively small number of essential genes across diverse bacteria (73). This is likely due to functional redundancy in addition to many genes contributing to bacterial fitness in untested habitats outside of the laboratory (47). Although previous transposon screens in P. syringae have provided information on traits required for epiphytic fitness and virulence, these have uncovered only those genes with large effects on behavior (74, 75). RB-TnSeq greatly expands our ability to interrogate the ecological determinants of such a cosmopolitan bacterium. Testing P. syringae and other bacterial species in a range of conditions, including those of ecological relevance, such as on and in additional host and nonhost plants, will enable the designation of functions for hypothetical or otherwise uncategorized proteins. Comparisons of these fitness assessments with specific in vitro experiments will enable the dissection of how individual genes contribute to a given process and to fitness on a eukaryotic host, a complex habitat with many distinct abiotic and biotic stressors. In such an approach, Cole et al. (43) used this method to examine specific nutrient requirements for P. simiae colonization of Arabidopsis roots. Many of the genes found to contribute to fitness had only small effects in planta. Expansion of these screens through additional generations of growth will increase the accumulated fitness defects, as seen in a recent study that sequentially passaged a Caulobacter crescentus transposon library to identify genes affecting attachment (76). Barcoded transposon libraries were originally developed as a highly scalable tool to identify gene function in diverse in vitro conditions, such as different growth conditions or abiotic stresses. Here we show that these same libraries can be used to better understand conditionally important genes that contribute to growth on the leaf surface and during colonization of the apoplast, expanding our understanding of the ecological fitness requirements on a genome-wide scale.
Materials and Methods
Bacterial Strains and Growth Media.
P. syringae pv. syringae B728a was originally isolated from a bean leaf (P. vulgaris) in Wisconsin (17). The complete genome for B728a is available on NCBI GenBank as accession no. CP000075.1 (77). B728a and derivative mutant strains were grown on KB agar or in broth (78) at 28 °C. E. coli strains S17-1, TOP10, and XL1-Blue were grown on LB agar or in LB broth at 37 °C. Geotrichum candidum Fr260 was grown at 25 °C on Potato Dextrose Agar (BD Difco). When appropriate, the following antibiotics were used at the indicated concentrations: 100 µg/mL rifampicin, 50 µg/mL kanamycin, 15 µg/mL tetracycline, 40 µg/mL nitrofurantoin, and 21.6 µg/mL natamycin.
Construction of Bar-Coded Transposon Library.
The bar-coded transposon insertion library was constructed by transposon mutagenesis using a bar-coded mariner transposon library, followed by TnSeq mapping and barcode association, as previously described (42). The E. coli WM3064 donor library containing the barcoded mariner plasmid, pKMW3, was recovered from a glycerol stock in LB kanamycin containing 300 µM diaminopimelic acid (DAP) and conjugated into B728a overnight on 50 different LB plates containing DAP. The many separate conjugations were then resuspended as a single pooled mixture and spread on 200 LB kanamycin plates for selecting mutants. Over 220,000 kanamycin-resistant B728a colonies were pooled for the library. All colonies were resuspended in 250 mL LB kanamycin and diluted to a starting OD600 0.2 for outgrowth at 28 °C with shaking to OD600 1.0. Finally, 250 µL 80% glycerol was added to 1-mL aliquots and frozen at −80 °C.
Plant Growth Conditions.
Common bean (P. vulgaris var. Blue Lake Bush 274) seeds (5 to 7 per 10-cm diameter pot) were planted in Super Soil (Scotts Miracle-Gro) and grown in a greenhouse for 2 wk before inoculation. Leaves were kept dry to minimize epiphytic bacterial populations. We used 1,000 W metal halide lights to provide supplemental lighting for a 16-h day length. Greenhouse temperatures ranged from 18 °C at night to as high as 40 °C during the day.
Library Recovery and Growth in KB.
For each inoculation, a 1.25 mL glycerol stock containing the transposon library was inoculated from −80 °C into 25 mL fresh KB with 100 µg/mL kanamycin and grown for ∼7 h at 28 °C with shaking until the culture reached midlog phase, OD600 0.5 to 0.7. Time0 samples were collected at this point during recovery; 1-mL aliquots were pelleted by centrifugation and the pellets were frozen until DNA purification. Cells were then washed twice in 10 mM KPO4 (pH = 7.0) prior to plant inoculation. To assay library growth in KB, 50-µL log phase cell culture (OD600 0.5) was inoculated into 950 µL KB with kanamycin in a 24-well plate. The plate was incubated at 28 °C with shaking for 15 h. Cells were collected by centrifugation, and frozen prior to DNA purification.
Plant Inoculations of the Transposon Library.
For epiphytic inoculations, cells were resuspended to a concentration of 2 × 106 CFU/mL in 10 mM KPO4 (OD600 = 0.001, by dilution from OD600 = 0.1), and sprayed onto the leaf surface until runoff; 100 pots were inoculated for each of 3 replicate experiments. Plants were then placed in a high-humidity chamber for 2 d.
For apoplastic inoculations, cells were resuspended to a concentration of 2 × 105 CFU/mL in 1 mM KPO4. The soil was covered with cotton to hold it in place, and the pots were inverted in ∼1.5 L inoculum in a glass bell jar. A vacuum was applied for 1.25 min and then removed rapidly to force the inoculum into the apoplast. Approximately 100 pots were inoculated for each of 3 replicate experiments. Plants were allowed to dry overnight and then moved to the greenhouse for 6 d.
Library Isolation from the Leaf Surface.
Leaves were collected and placed in a glass dish filled with dH2O, which was placed in a sonication water bath (Branson 5510, output frequency 40 kHz), and sonicated for 10 min to remove cells. Approximately 1 L dH2O + leaves was sonicated at a time. The resulting leaf wash was filtered through a 6-µm filter (Whatman #3), and then cells were collected on 0.2-µm filters. Cells were removed from the filters by vortexing in 30 mL total 10 mM KPO4, and centrifuged at 17,000 × g for 1 min to pellet. Cell pellet aliquots were frozen prior to DNA purification.
Library Isolation from the Apoplast.
Leaves were chopped in a blender to yield fragments of about 2 to 3 mm in diameter, and placed in a glass dish filled with dH2O, placed in a sonication water bath, and sonicated for 15 min to remove cells. Leaves were not surface-sterilized to enable leaf sampling at this scale. Since the number of epiphytic cells on leaves kept at low humidity was several orders-of-magnitude lower than those in the apoplast the contribution of any epiphytic cells to the recovered apoplastic cells was negligible. Approximately 1 L leaf slurry was sonicated at a time. The resulting slurry was filtered through a coffee filter to minimize plant debris. Ten percent of the ∼5- to 10-L buffer was taken for additional filtration steps. This sample was filtered through several Whatman filters (20 µm, 10 µm, and 6 µm), and then concentrated by centrifugation at 4,696 × g for 10 min. The pellet was resuspended in 10 mM KPO4, and aliquots of cell pellets were frozen prior to DNA purification.
DNA Isolation and Library Preparation.
DNA from frozen pellets was isolated using the Qiagen DNeasy Blood & Tissue Kit according to the manufacturer’s instructions. Cell lysis was done at 50 °C for 10 min as per optional instructions. For apoplastic samples with excess plant material, lysed cells were centrifuged at 1,500 × g for 5 min before loading the supernatant onto Qiagen purification columns. Purified genomic DNA was measured on a nanodrop and 200 ng of total DNA was used as a template for DNA barcode amplification and adapter ligation, as established previously (42). For each time0 and plant experimental sample, 2 separately purified DNA samples were sequenced as technical replicates.
Sequencing and Fitness Data Generation.
Barcode sequencing, mapping, and analysis to calculate the relative abundance of barcodes was done using the RB-TnSeq methodology and computation pipeline developed by Wetmore et al. (42); the code is available at https://bitbucket.org/berkeleylab/feba/. TnSeq was used to map the insertion sites and associate the DNA barcodes to these insertions. Based on the TnSeq data, standard computational methods (47) were used to predict which genes are likely essential for viability in LB. For these data, the minimum gene length to call a gene essential was 325 bp. For each experiment, fitness values for each gene are calculated as a log2 ratio of relative barcode abundance following library growth in a given condition divided by relative abundance in the time0 sample. Fitness values are normalized across the genome so the typical gene has a fitness value of 0. All experiments passed previously described quality-control metrics (42), with the exception of 1 technical replicate of apoplast_2, which was removed from our analysis. Fitness values from sequencing replicates were averaged for each experiment. Experimental fitness values are publicly available at fit.genomics.lbl.gov (79). Additional data, including BarSeq counts for all insertional strains, are publicly available at https://github.com/thelmann/P.syringae-leaf-colonization (80).
Comparison of Predicted Essential Genes in B728a to Other Pseudomonas Species.
We used the IMG database (44) to identify homologs for B728a genes in P. aeruginosa PAO1, P. stutzeri RCH2, and P. simiae WCS417 using the genome-gene best-homologs function at 70% identity. Turner et al. (46) predicted 336 essential genes in P. aeruginosa PAO1 using a Monte Carlo statistical analysis. Essential genes in P. stutzeri RCH2 and P. simiae WCS417 were predicted previously (47). A comparison of B728a genes predicted to be essential or nearly essential (n = 392) with their orthologs in these strains identified 3 categories: Predicted essential and nonessential genes, and B728a genes with no orthologs identified.
Genomic Fitness Data Analysis.
A dendrogram of experiments was generated from the matrix of fitness values using the dist and hclust functions in R (81) with the default clustering method “Euclidean.” To better classify genes based on their genomic annotation, we assigned gene names, gene product descriptions, and broad functional categories based on the previously annotated genomic metadata (27). For each gene, fitness values for experimental replicates were averaged to calculate an average gene fitness value for each treatment. We focused our analysis on genes with average fitness less than −2 and t < −3 in at least 2 experimental replicates. However, we also considered genes for analysis with average fitness less than −1 and t < −2.5 in at least 2 experimental replicates. The t-score is a test statistic used to assess the statistical significance of the gene fitness value (42). For each functional category, we used a the phyper function in R to perform a hypergeometric test to examine category enrichment, using average fitness less than −2. Graphs were plotted in R using the ggplot2 package, v3.1.1 (82). Heatmaps were plotted in R using the function heatmap.2 in the gplots package, v3.0.1.1 (83).
Construction of Targeted Deletion Mutants.
Deletion strains were constructed using an overlap extension PCR protocol as described previously (84). Briefly, 1-kb DNA fragments upstream and downstream of the genes of interest were amplified along with a kanamycin-resistance cassette from pKD13 (85). These 3 fragments were joined by overlap extension PCR. The resulting fragment was blunt-end ligated into the SmaI site of pTsacB (86), and transformed into the E. coli subcloning strains TOP10 or XL1-Blue, and then the E. coli conjugation donor strain S17-1. This suicide plasmid was conjugated into B728a on KB overnight, and then selected for 3 d on KB containing kanamycin and nitrofurantoin (E. coli counter selection). Putative double-crossover colonies that were kanamycin-resistant and tetracycline-sensitive were selected for screening using external primers and insertion sites further confirmed by PCR and Sanger sequencing.
Bacterial In Vitro Growth Measurements.
Strains were grown overnight in KB broth with rifampicin, washed 3 times in 10 mM KPO4, and standardized to OD600 = 0.3. Twenty microliters was inoculated per well into a 96-well plate containing 100 µL KB broth, LB broth, or M9 broth supplemented with 0.2% (vol/vol) glycerol (3 to 5 replicate wells for each strain). Auxotrophic mutant strains were functionally complemented by the addition of either 20.8 µg/mL tryptophan or 62 µg/mL histidine. The plate was sealed using a semipermeable membrane (Breathe-Easy Sealing Film, Diversified Biotech). Cells were grown at 28 °C with shaking, taking absorbance measurements at 600 nm every 30 min. Absorbance values are reported, with the average absorbance for 2 replicate media blanks subtracted from the total absorbance for each well.
Bioassay of the Syringomycin Mutant Strain.
G. candidum was grown at 25 °C for 2 d on a potato dextrose agar (PDA) plate. Bacterial strains were grown overnight in KB with rifampicin, washed in 10 mM KPO4, and standardized to OD600 = 0.3. A 10-µL suspension was spotted onto PDA plates, and grown at 25 °C for 3 d. After incubation, the plates were lightly oversprayed with a dense fungal suspension and zones of fungal growth inhibition were measured after 24 h.
Bacterial Apoplastic Growth Measurements.
Strains were grown overnight on KB with rifampicin, washed in 10 mM KPO4, and standardized to 2 × 105 CFU/mL in 1 mM KPO4. Cells were inoculated into leaves of 2-wk-old plants using a blunt syringe. Leaf samples (3 discs per leaf) were taken using a 5-mm-diameter cork borer into tubes containing 200 µL 10 mM KPO4 and 2 3-mm glass beads, and ground for 30 s at 2,400 rpm in a Mini-Beadbeater-96 (Biospec Products) before dilution plating on KB with rifampicin and natamycin.
Comparison of Gene Expression and Contribution to Fitness.
B728a gene expression was previously determined as a measure of microarray fluorescence from RNA isolated from cells in hrp-inducing MM, epiphytic growth, or apoplastic growth in P. vulgaris (27). These data were originally generated by 2 laboratories, and within-laboratory technical replicates were averaged before calculating the mean fluorescence for epiphytic and apoplastic treatments. Fold-change was calculated as a log2 of the ratio of epiphytic or apoplastic fluorescence divided by fluorescence in MM. Gene expression for strain DC3000 in the apoplast of A. thaliana and in KB was previously measured by RNA sequencing. (28). Fold-change for a given gene was calculated as a log2 of the ratio of apoplastic reads per million divided by reads per million in KB. Orthologs for 4,209 B728a genes were identified in strain DC3000 using the IMG genome-gene best-homologs function at 70% identity (44). Of these, fitness values could be calculated for 1,925 genes in B728a for which RNA-seq expression patterns were available in DC3000, and are plotted here.
Supplementary Material
Acknowledgments
We thank Morgan Price for assistance with RB-TnSeq sequence analysis; Jayashree Ray for mapping the insertion sites of the B728a transposon library; Russell Scott for the B728a ∆hrpL strain; Dennis Gross for the Geotrichum candidum isolate; and Dana King, Caitlin Ongsarte, and Jennifer Lam for assistance with plant inoculations. Funding for T.C.H. was partially provided by the Arnon Graduate Fellowship and the William Carroll Smith Fellowship. This work used the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley, supported by NIH S10 OD018174 Instrumentation Grant.
Footnotes
The authors declare no conflict of interest.
Data deposition: Fitness values from sequencing replicates were averaged for each experiment. Experimental fitness values are publicly available at fit.genomics.lbl.gov. Additional data, including BarSeq counts for all insertional strains, are publicly available at https://github.com/thelmann/P.syringae-leaf-colonization.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908858116/-/DCSupplemental.
References
- 1.Lindemann J., Arny D. C., Upper C. D., Use of an apparent infection threshold population of Pseudomonas syringae to predict incidence and severity of brown spot of bean. Phytopathology 74, 1334–1339 (1984). [Google Scholar]
- 2.Hirano S. S., Upper C. D., Ecology and epidemiology of foliar bacterial plant pathogens. Annu. Rev. Phytopathol. 21, 243–269 (1983). [Google Scholar]
- 3.Melotto M., Underwood W., He S. Y., Role of stomata in plant innate immunity and foliar bacterial diseases. Annu. Rev. Phytopathol. 46, 101–122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leveau J. H. J., Lindow S. E., Appetite of an epiphyte: Quantitative monitoring of bacterial sugar consumption in the phyllosphere. Proc. Natl. Acad. Sci. U.S.A. 98, 3446–3453 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Wal A., Leveau J. H. J., Modelling sugar diffusion across plant leaf cuticles: The effect of free water on substrate availability to phyllosphere bacteria. Environ. Microbiol. 13, 792–797 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Mercier J., Lindow S. E., Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl. Environ. Microbiol. 66, 369–374 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vorholt J. A., Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Lindow S. E., Brandl M. T., Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Leary B. M., et al. , Early changes in apoplast composition associated with defence and disease in interactions between Phaseolus vulgaris and the halo blight pathogen Pseudomonas syringae Pv. phaseolicola. Plant Cell Environ. 39, 2172–2184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beattie G. A., Water relations in the interaction of foliar bacterial pathogens with plants. Annu. Rev. Phytopathol. 49, 533–555 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Aung K., Jiang Y., He S. Y., The role of water in plant-microbe interactions. Plant J. 93, 771–780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xin X. F., et al. , Bacteria establish an aqueous living space in plants crucial for virulence. Nature 539, 524–529 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin X. F., Kvitko B., He S. Y., Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindemann J., Arny D. C., Upper C. D., Epiphytic populations of Pseudomonas syringae pv. syringae on snap bean and nonhost plants and the incidence of bacterial brown spot disease in relation to cropping patterns. Ecol. Epidemiol. 74, 1329–1333 (1984). [Google Scholar]
- 15.Morris C. E., et al. , The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2, 321–334 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Monteil C. L., Bardin M., Morris C. E., Features of air masses associated with the deposition of Pseudomonas syringae and Botrytis cinerea by rain and snowfall. ISME J. 8, 2290–2304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loper J. E., Lindow S. E., Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77, 1449–1454 (1987). [Google Scholar]
- 18.Baltrus D. A., McCann H. C., Guttman D. S., Evolution, genomics and epidemiology of Pseudomonas syringae. Mol. Plant Pathol. 18, 152–168 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris C. E., et al. , Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe, and New Zealand. MBio 1, e00107-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinatzer B. A., et al. , The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 62, 26–44 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Morris C. E., Lamichhane J. R., Nikolić I., Stanković S., Moury B., The overlapping continuum of host range among strains in the Pseudomonas syringae complex. Phytopathol Res. 1, 1–16 (2019). [Google Scholar]
- 22.Lindow S. E., The role of bacterial ice nucleation in frost injury to plants. Annu. Rev. Phytopathol. 21, 363–384 (1983). [Google Scholar]
- 23.Baltrus D. A., et al. , Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7, e1002132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bender C. L., Alarcón-Chaidez F., Gross D. C., Pseudomonas syringae phytotoxins: Mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63, 266–292 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris C. E., Monteil C. L., Berge O., The life history of Pseudomonas syringae: Linking agriculture to earth system processes. Annu. Rev. Phytopathol. 51, 85–104 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Gal M., Preston G. M., Massey R. C., Spiers A. J., Rainey P. B., Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol. Ecol. 12, 3109–3121 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Yu X., et al. , Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. U.S.A. 110, E425–E434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobori T., et al. , Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. U.S.A. 115, E3055–E3064 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindgren P. B., et al. , An ice nucleation reporter gene system: Identification of inducible pathogenicity genes in Pseudomonas syringae pv. phaseolicola. EMBO J. 8, 1291–1301 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boch J., et al. , Identification of Pseudomonas syringae pv. tomato genes induced during infection of Arabidopsis thaliana. Mol. Microbiol. 44, 73–88 (2002). [DOI] [PubMed] [Google Scholar]
- 31.Marco M. L., Legac J., Lindow S. E., Pseudomonas syringae genes induced during colonization of leaf surfaces. Environ. Microbiol. 7, 1379–1391 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Kinkel L. L., Wilson M., Lindow S. E., Effect of sampling scale on the assessment of epiphytic bacterial populations. Microb. Ecol. 29, 283–297 (1995). [DOI] [PubMed] [Google Scholar]
- 33.Price M. N., et al. , Indirect and suboptimal control of gene expression is widespread in bacteria. Mol. Syst. Biol. 9, 660 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X. X., George A., Bailey M. J., Rainey P. B., The histidine utilization (hut) genes of Pseudomonas fluorescens SBW25 are active on plant surfaces, but are not required for competitive colonization of sugar beet seedlings. Microbiology 152, 1867–1875 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Turner K. H., Everett J., Trivedi U., Rumbaugh K. P., Whiteley M., Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 10, e1004518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McAdam P. R., Richardson E. J., Fitzgerald J. R., High-throughput sequencing for the study of bacterial pathogen biology. Curr. Opin. Microbiol. 19, 106–113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rastogi G., Coaker G. L., Leveau J. H. J., New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 348, 1–10 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Van Sluys M. A., et al. , Comparative genomic analysis of plant-associated bacteria. Annu. Rev. Phytopathol. 40, 169–189 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Levy A., et al. , Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melnyk R. A., Hossain S. S., Haney C. H., Convergent gain and loss of genomic islands drive lifestyle changes in plant-associated Pseudomonas. ISME J. 13, 1575–1588 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Opijnen T., Bodi K. L., Camilli A., Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wetmore K. M., et al. , Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 6, e00306–e00315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cole B. J., et al. , Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 15, e2002860 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen I. A., et al. , IMG/M v.5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacobs M. A., et al. , Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 100, 14339–14344 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Turner K. H., Wessel A. K., Palmer G. C., Murray J. L., Whiteley M., Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl. Acad. Sci. U.S.A. 112, 4110–4115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Price M. N., et al. , Mutant phenotypes for thousands of bacterial genes of unknown function. Nature 557, 503–509 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Andersen G. L., Beattie G. A., Lindow S. E., Molecular characterization and sequence of a methionine biosynthetic locus from Pseudomonas syringae. J. Bacteriol. 180, 4497–4507 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris J. J., Lenski R. E., Zinser E. R., The Black Queen Hypothesis: Evolution of dependencies through adaptive gene loss. MBio 3, e00036-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S. A., et al. , General and condition-specific essential functions of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 5189–5194 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beattie G. A., Lindow S. E., Survival, growth, and localization of epiphytic fitness mutants of Pseudomonas syringae on leaves. Appl. Environ. Microbiol. 60, 3790–3798 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rico A., Preston G. M., Pseudomonas syringae pv. tomato DC3000 uses constitutive and apoplast-induced nutrient assimilation pathways to catabolize nutrients that are abundant in the tomato apoplast. Mol. Plant Microbe Interact. 21, 269–282 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Duong D. A., Jensen R. V., Stevens A. M., Discovery of Pantoea stewartii ssp. stewartii genes important for survival in corn xylem through a Tn-Seq analysis. Mol. Plant Pathol. 19, 1929–1941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Royet K., Parisot N., Rodrigue A., Gueguen E., Condemine G., Identification by Tn-seq of Dickeya dadantii genes required for survival in chicory plants. Mol. Plant Pathol. 20, 287–306 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Erbs G., Newman M. A., The role of lipopolysaccharide and peptidoglycan, two glycosylated bacterial microbe-associated molecular patterns (MAMPs), in plant innate immunity. Mol. Plant Pathol. 13, 95–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rocchetta H. L., Burrows L. L., Lam J. S., Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 63, 523–553 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rapicavoli J. N., et al. , Lipopolysaccharide O-antigen delays plant innate immune recognition of Xylella fastidiosa. Nat. Commun. 9, 390 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeuchi K., et al. , Flagellin glycosylation island in Pseudomonas syringae pv. glycinea and its role in host specificity. J. Bacteriol. 185, 6658–6665 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taguchi F., et al. , Identification of glycosylation genes and glycosylated amino acids of flagellin in Pseudomonas syringae pv. tabaci. Cell. Microbiol. 8, 923–938 (2006). [DOI] [PubMed] [Google Scholar]
- 60.Buscaill P., et al. , Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364, eaav0748 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Yu J., Peñaloza-Vázquez A., Chakrabarty A. M., Bender C. L., Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol. Microbiol. 33, 712–720 (1999). [DOI] [PubMed] [Google Scholar]
- 62.Markel E., Stodghill P., Bao Z., Myers C. R., Swingle B., AlgU controls expression of virulence genes in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 198, 2330–2344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peñaloza-Vázquez A., Kidambi S. P., Chakrabarty A. M., Bender C. L., Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. J. Bacteriol. 179, 4464–4472 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu X., et al. , Transcriptional analysis of the global regulatory networks active in Pseudomonas syringae during leaf colonization. MBio 5, e01683-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang W. S., et al. , Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J. Bacteriol. 189, 8290–8299 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kvitko B. H., et al. , Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5, e1000388 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu G. W., Gross D. C., Evaluation of the role of syringomycin in plant pathogenesis by using Tn5 mutants of Pseudomonas syringae pv. syringae defective in syringomycin production. Appl. Environ. Microbiol. 54, 1345–1353 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hulin M. T., et al. , Comparative genomics of Pseudomonas syringae reveals convergent gene gain and loss associated with specialization onto cherry (Prunus avium). New Phytol. 219, 672–696 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Schellenberg B., Ramel C., Dudler R., Pseudomonas syringae virulence factor syringolin A counteracts stomatal immunity by proteasome inhibition. Mol. Plant Microbe Interact. 23, 1287–1293 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Scholz-Schroeder B. K., Hutchison M. L., Grgurina I., Gross D. C., The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant Microbe Interact. 14, 336–348 (2001). [DOI] [PubMed] [Google Scholar]
- 71.Hutchison M. L., Gross D. C., Lipopeptide phytotoxins produced by Pseudomonas syringae pv. syringae: Comparison of the biosurfactant and ion channel-forming activities of syringopeptin and syringomycin. Mol. Plant Microbe Interact. 10, 347–354 (1997). [DOI] [PubMed] [Google Scholar]
- 72.Huse H. K., et al. , Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. MBio 1, e00199-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luo H., Lin Y., Gao F., Zhang C. T., Zhang R., DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 42, D574–D580 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manoharan B., Neale H. C., Hancock J. T., Jackson R. W., Arnold D. L., The identification of genes important in Pseudomonas syringae pv. phaseolicola plant colonisation using in vitro screening of transposon libraries. PLoS One 10, e0137355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindow S. E., Andersen G., Beattie G. A., Characteristics of insertional mutants of Pseudomonas syringae with reduced epiphytic fitness. Appl. Environ. Microbiol. 59, 1593–1601 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hershey D. M., Fiebig A., Crosson S., A genome-wide analysis of adhesion in Caulobacter crescentus identifies new regulatory and biosynthetic components for holdfast assembly. MBio 10, e02273-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feil H., et al. , Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U.S.A. 102, 11064–11069 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.King E. O., Ward M. K., Raney D. E., Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307 (1954). [PubMed] [Google Scholar]
- 79.Price M., Lo V., Shao W., Gammaproteobacteria: Pseudomonas syringae pv. syringae B728a. The Fitness Browser. http://fit.genomics.lbl.gov/cgi-bin/org.cgi?orgId=SyringaeB728a. Deposited 27 June 2019.
- 80.Helmann T., BarSeq all.poolcount. GitHub. https://github.com/thelmann/P.syringae-leaf-colonization. Deposited 29 June 2019.
- 81.R Core Team , R: A Language and Environment for Statistical Computing (Version 3.4.3, R Foundation for Statistical Computing, Vienna, Austria, 2017).
- 82.Wickham H., ggplot2: Elegant Graphics for Data Analysis (Version 3.1.1, Springer, New York, 2016). https://ggplot2.tidyverse.org. Accessed 1 May 2019.
- 83.Warnes G. R., et al. , gplots: Various R Programming Tools for Plotting Data (Version 3.0.1.1, 2019). https://cran.r-project.org/package=gplots. Accessed 1 May 2019.
- 84.Hockett K. L., Burch A. Y., Lindow S. E., Thermo-regulation of genes mediating motility and plant interactions in Pseudomonas syringae. PLoS One 8, e59850 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Datsenko K. A., Wanner B. L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen C., Malek A. A., Wargo M. J., Hogan D. A., Beattie G. A., The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75, 29–45 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.